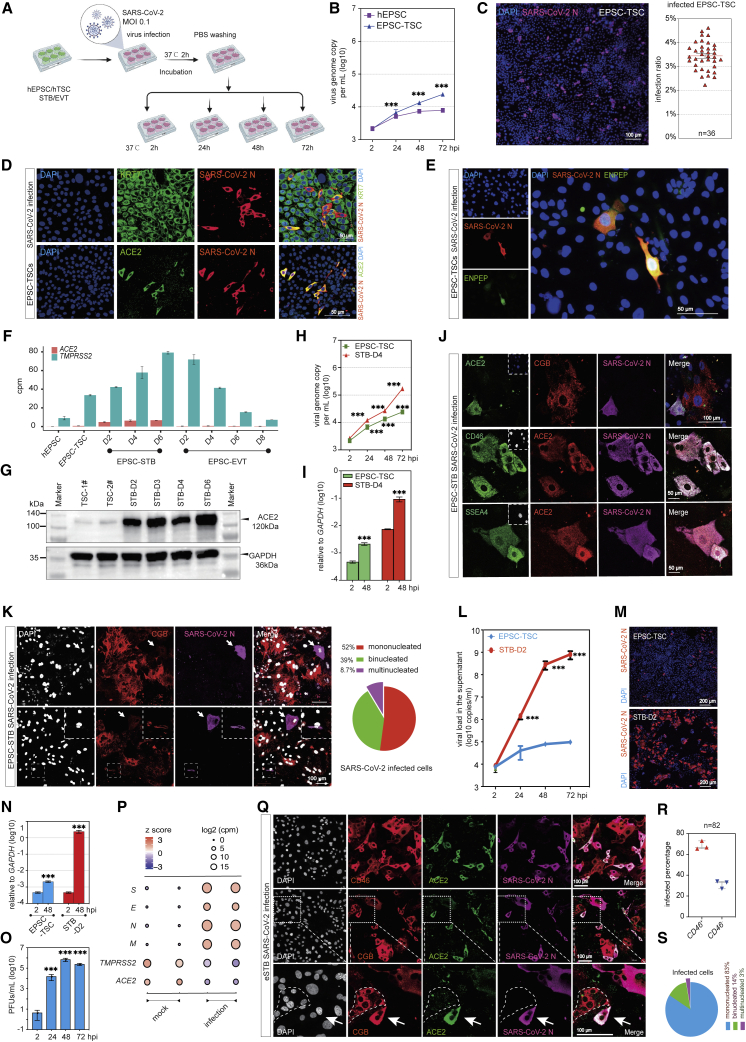
Figure 3.
eSTBs are highly susceptible to SARS-CoV-2 infection among trophoblasts
(A) Schematic diagram of SARS-CoV-2 infection of in vitro trophoblasts.
(B) qRT-PCR detection of SARS-CoV-2 genome copy numbers (per mL) in the supernatants of virus-co-cultured cells at different time points. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗∗∗p < 0.001.
(C) Immunofluorescence image of 24 hpi EPSC-TSCs stained for SARS-CoV-2 N protein. Scale bar, 100 μm. Right: percentages of N-protein-positive cells. Data are mean ± SEM; n = 36, quantification of 36 random images.
(D) Immunofluorescence detection of KRT7, ACE2, and N protein in the infected EPSC-TSCs. Scale bars, 50 μm.
(E) Representative immunofluorescence images of N protein and ENPEP at 24 hpi in EPSC-TSCs. Scale bar, 50 μm.
(F) Bar plot for expression (cpm) of ACE2 and TMPRSS2 in hEPSCs, EPSC-TSCs, and STBs and EVTs differentiated from EPSC-TSCs at indicated time points.
(G) Detection of ACE2 protein in EPSC-TSCs and during their differentiation toward STBs.
(H) qRT-PCR detection of SARS-CoV-2 genome copy numbers (per mL) in the supernatants of virus-co-cultured EPSC-TSCs and (day 4) STBs. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(I) qRT-PCR analysis of SARS-CoV-2 genome copy number in cell lysates of 48 hpi EPSC-TSCs and STB-D4. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(J) Immunofluorescence staining of 48 hpi STB-D6 for ACE2, CGB, and N protein and for early STB markers CD46 and SSEA4. Scale bars, 50 or 100 μm as specified.
(K) Immunofluorescence staining of 48 hpi STB-D6 for ACE2, CGB, and N protein, and percentages of mono-, bi-, and multinucleated CGB+ cells in the infected STB-D6. Arrowheads indicate that some CGB-low or mono- or binucleated cells were infected by SARS-CoV-2. DAPI stains the nucleus. Scale bars, 100 μm.
(L) qRT-PCR detection of SARS-CoV-2 genome copy number (per mL) in the supernatants of virus-co-cultured EPSC-TSCs and eSTBs (STB-D2). Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(M) Representative immunofluorescence images of 24 hpi EPSC-TSCs and eSTBs (STB-D2) stained for N protein. Scale bars, 200 μm.
(N) qRT-PCR detection of SARS-CoV-2 gene expression in cell lysates of the infected cells at 48 hpi. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(O) Plaque formation assay to detect SARS-CoV-2 virus in supernatants from the infected eSTBs (48 hpi). Data are mean ± SD; n = 3 three independent experiments; ∗∗∗p < 0.001 (Student’s t test). PFUs, plaque-forming units.
(P) Bubble plot for RNA-seq analysis of SARS-CoV-2 genes (E, M, N, S) in the cell lysates of 48 hpi eSTBs.
(Q) Representative immunofluorescence images of 24 hpi eSTBs stained for ACE2 and N protein and for STB markers CD46 and CGB. The dotted area shows an uninfected multinucleated CGB+ cell. Arrows indicate an infected binucleated STB. Scale bars, 100 μm.
(R) Quantification of the percentages of eSTB marker CD46 in 24 hpi eSTBs. Data are mean ± SEM.
(S) Quantification of the percentages of mono-, bi-, and multinucleated cells in 24 hpi eSTBs. SARS-CoV-2 preferentially infects mononucleated cells.