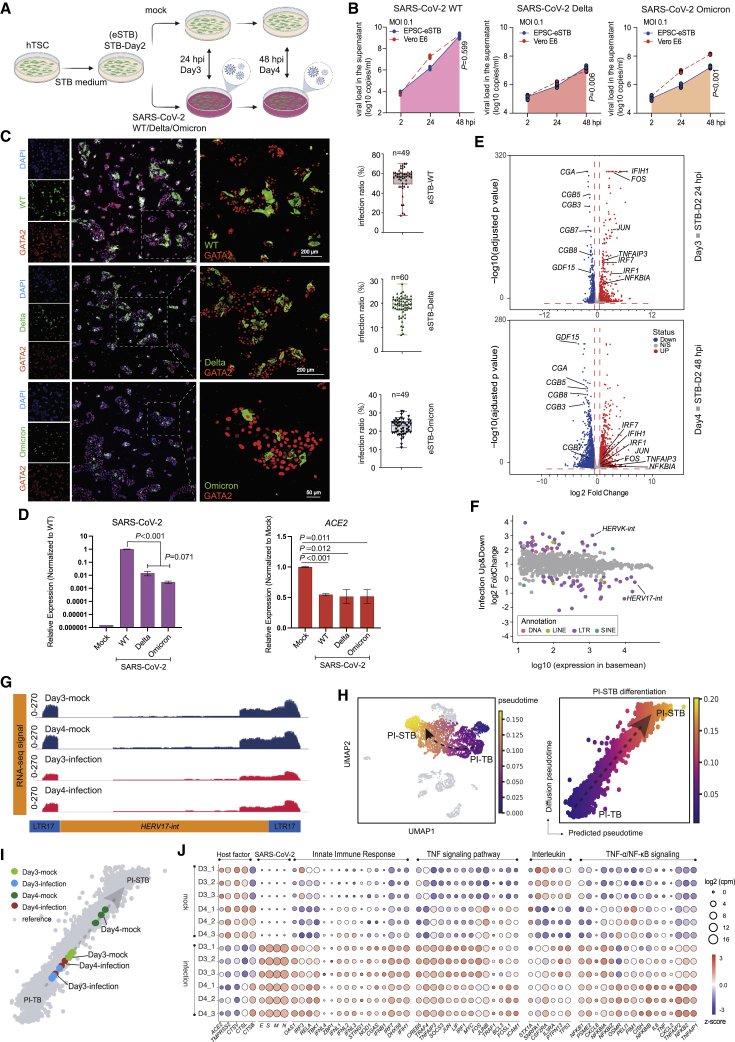
Figure 4.
SARS-CoV-2 variants robustly replicate in eSTBs
(A) Schematic diagram of eSTBs for SARS-CoV-2 wild-type (WT) and Delta or Omicron variant infection.
(B) Comparison of SARS-CoV-2 replication kinetics in EPSC-eSTBs and Vero E6 cells.
(C) Left: representative immunofluorescence images of N protein and GATA2 in EPSC-eSTBs infected by SARS-CoV-2 WT, Delta, and Omicron at 24 hpi. Right: percentages of N protein-positive EPSC-eSTBs. Error bars: mean and SEM; n = 49, 60, and 49 for quantification of random immunofluorescence images in WT-, Delta-, and Omicron-infected eSTBs, respectively.
(D) qRT-PCR analysis of ACE2 and virus gene expression levels in 48 hpi EPSC-eSTBs. Data are mean ± SD; n = 3. Student’s t test, exact p values are presented.
(E) Volcano plots of gene expression in infected STB-D3 and STB-D4. The horizontal red dashed line marks the adjusted p value (Wald test) 0.05, while the vertical red dashed lines marks expression fold change of 1.5.
(F) A scatter diagram of the transcriptomic analysis of TEs after virus infection. Differentially expressed TEs (p < 0.05) are in colors, while non-differentially expressed TEs are in gray. Expression levels of TEs used for the x axis are from DESeq2 results. Data from STB-D3 and STB-D4 are combined in this analysis.
(G) RNA-seq signal of HERV-W in infected STB-3 and STB-4 and the mock-infection control cells. Library size was used to normalize the read counts.
(H) Pseudotime analysis depicting PI-TB to PI-STB development trajectory. The black dashed arrow indicates the imputed direction of differentiation. PI-TB to PI-STB subpopulations are colored by machine learning predicted pseudotime. The x axis is the predicted pseudotime, and the y axis is the diffusion pseudotime computed using SCANPY. Gray dashed arrow describes the linear regression relationship between the predicted pseudotime and the diffusion pseudotime.
(I) Infected vs. normal (mock) STB-D3 and STB-D4 mapped against the PI-TB to PI-STB pseudotime trajectory (see STAR Methods).
(J) Bubble plot for host factors ACE2, TMPRSS2, CTSL, CTSV, and CTSB; infection-related signaling pathways; and virus genes in infected eSTBs. Bubble colors are in accordance with Z score and bubble sizes are proportional to expression levels.