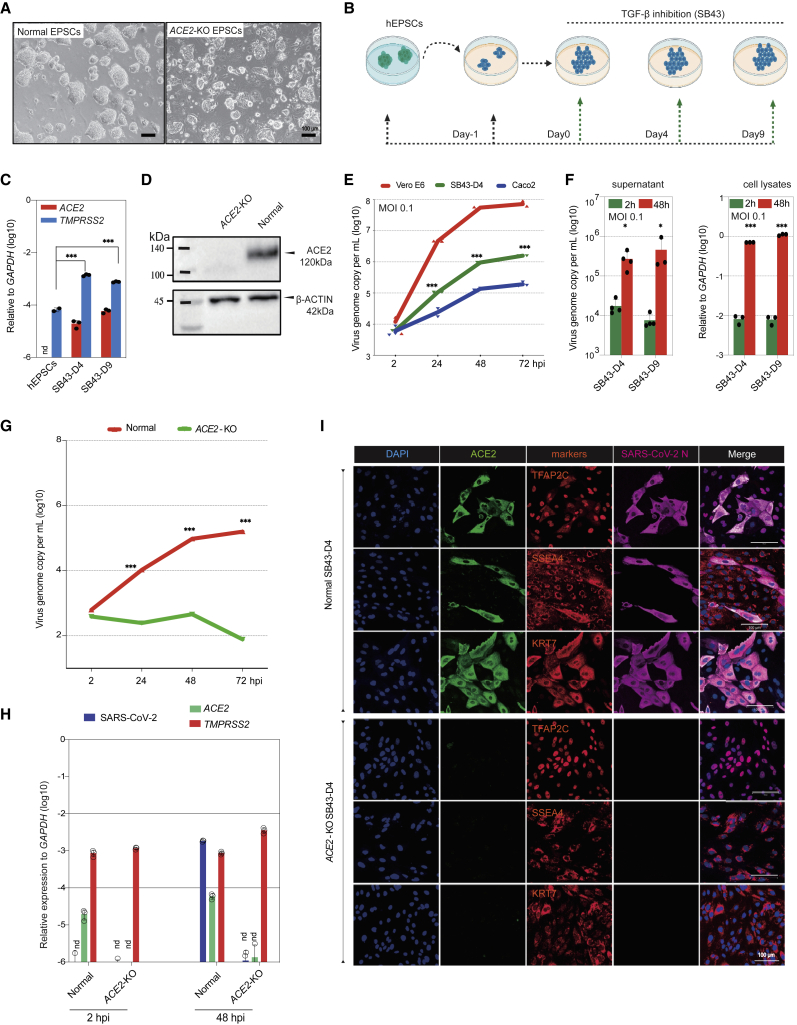
Figure 7.
ACE2 is required for SARS-CoV-2 infection of trophoblasts differentiated from hEPSCs
(A) Normal and ACE2-KO hEPSCs. Scale bars, 100 μm.
(B) Experimental flow of hEPSC differentiation toward trophoblasts via TGF-β inhibitor SB-431542 (SB43) treatment.
(C) qRT-PCR analysis of ACE2 and TMPRSS2 in hEPSCs and the differentiated cells on days 4 and 9. nd, undetectable. Data are mean ± SD; n = 3; ∗∗∗p < 0.001 (Student’s t test).
(D) Western blotting confirms loss of ACE2 protein in the SB43-treated hEPSCs on day 9 (SB43D9) of differentiation.
(E) SARS-CoV-2 replication kinetics in cells differentiated from hEPSCs compared with Vero E6 and Caco2 cells. Data are mean ± SD; n = 3 biological replicates; ∗∗∗p < 0.001 (Student’s t test).
(F) Left: qRT-PCR of supernatant viral load in SB43-treated hEPSCs at 2 and 48 hpi. Right: qRT-PCR quantification of SARS-CoV-2 genome in cell lysates of SB43-treated hEPSCs on days 4 and 9. Data are mean ± SD; n = 3 biological replicates; ∗p < 0.05, ∗∗∗p < 0.001 (Student’s t test).
(G) Quantification of supernatant viral RNA loads of SB43-treated (day 4) normal and ACE2-KO EPSCs. Data are mean ± SD; n = 3 biological replicates; ∗∗∗p < 0.001 (Student’s t test).
(H) SARS-CoV-2 viral genome quantitation and expression of ACE2 and TMPRSS2 in normal vs. SB43-treated ACE2-KO EPSCs at day 4. Data are mean ± SD; n = 3 biological replicates; nd, not detectable.
(I) Representative immunofluorescence staining images for ACE2, trophoblast factors and markers, and SARS-CoV-2 N protein in normal (top) and ACE2-KO cells (bottom) (day 4 of SB43 treatment) at 48 hpi. Scale bars, 100 μm.