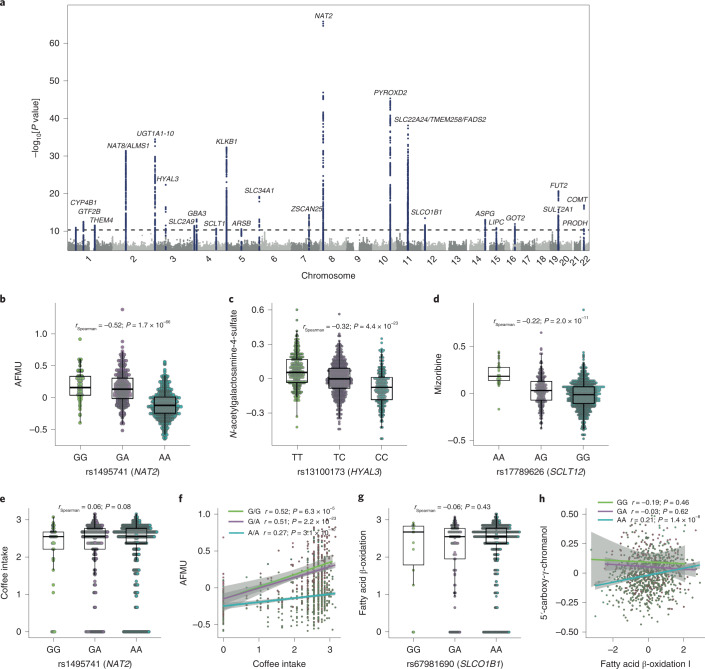
Fig. 4. Genetic associations of plasma metabolites.
a, Manhattan plot showing 48 independent mQTLs identified linking 44 metabolites and 40 genetic variants with P < 4.2 × 10−11 (Spearman; two sided). Representative genes for the SNPs with significant mQTLs are labeled. b, Association between a tag SNP (rs1495741) of the NAT2 gene and plasma AFMU levels. c, Association between a SNP (rs13100173) within the HYAL3 gene and plasma levels of N-acetylgalactosamine-4-sulfate. d, Association between a tag SNP (rs17789626) of the SCLT1 gene and plasma mizoribine levels. e, Differences in coffee intake between participants with different genotypes at rs1495741. f, Correlations between coffee intake and AFMU in participants with different genotypes at rs1495741. g, Differences in bacterial fatty acid β-oxidation pathway abundance in participants with different genotypes at rs67981690. h, Correlations between bacterial fatty acid β-oxidation pathway abundance and 5′-carboxy-γ-chromanol in participants with different genotypes at rs67981690. In b–e and g, the x axis indicates the genotype of the corresponding SNP and the y axis indicates normalized residuals of the corresponding metabolic abundance (n = 927 biologically independent samples). Each dot represents one sample. The box plots show the median and first and third quartiles (25th and 75th percentiles) of the metabolite levels. The upper and lower whiskers extend to the largest and smallest value no further than 1.5× the IQR, respectively. Outliers are plotted individually. The association strength is shown by the Spearman correlation coefficient and corresponding P value (two sided). In f and h, the x axis indicates the normalized abundance of coffee intake (f) or the bacterial fatty acid β-oxidation pathway (h) and the y axis indicates the normalized residuals of the corresponding metabolic abundance. Each dot represents one sample (n = 927 biologically independent samples). The lines indicate linear regressions for each genotype group separately. Areas with light gray shading indicate the 95% CI of the linear regression lines. The association strength per genotype is shown by the Spearman correlation and the corresponding P value (two sided).