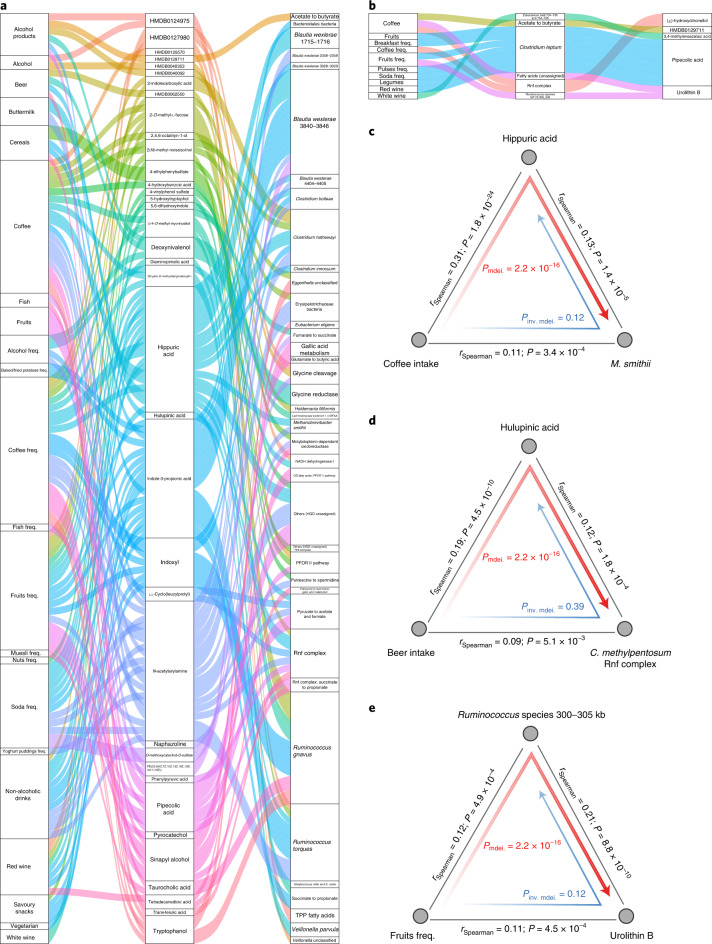
Fig. 6. Mediation analysis identifies linkages between the gut microbiome, metabolites and dietary habits.
a, Parallel coordinates chart showing the 133 mediation effects of plasma metabolites that were significant at FDR < 0.05. Shown are dietary habits (left), plasma metabolites (middle) and microbial factors (right). The curved lines connecting the panels indicate the mediation effects, with colors corresponding to different metabolites. freq., frequency; PFOR, pyruvate:ferredoxin oxidoreductase; OD, oxidative decarboxylation; HGD, 2-hydroxyglutaryl-CoA dehydratase; TPP, thiamine pyrophosphate. b, Parallel coordinates chart showing the 13 mediation effects of the microbiome that were significant at FDR < 0.05. Shown are dietary habits (left), microbial factors (middle) and plasma metabolites (right). For the microbial factors column, number ranges represent the genomic location of microbial structure variations (SVs) in kilobyte unit, and colons represent the detailed annotation of certain gutSMASH pathway. c, Analysis of the effect of coffee intake on the abundance of M. smithii as mediated by hippuric acid. d, Analysis of the effect of beer intake on the C. methylpentosum Rnf complex pathway as mediated by hulupinic acid. e, Analysis of the effect of fruit intake on urolithin B in plasma as mediated by a vSV in Ruminococcus species (300‒305 kb). In c–e, the gray lines indicate the associations between the two factors, with corresponding Spearman coefficients and P values (two sided). Direct mediation is shown by a red arrow and reverse mediation is shown by a blue arrow. Corresponding P values from mediation analysis (two sided) are shown. inv., inverse; mdei., mediation.