Figure 2. Single cell sequencing of mtDNA.
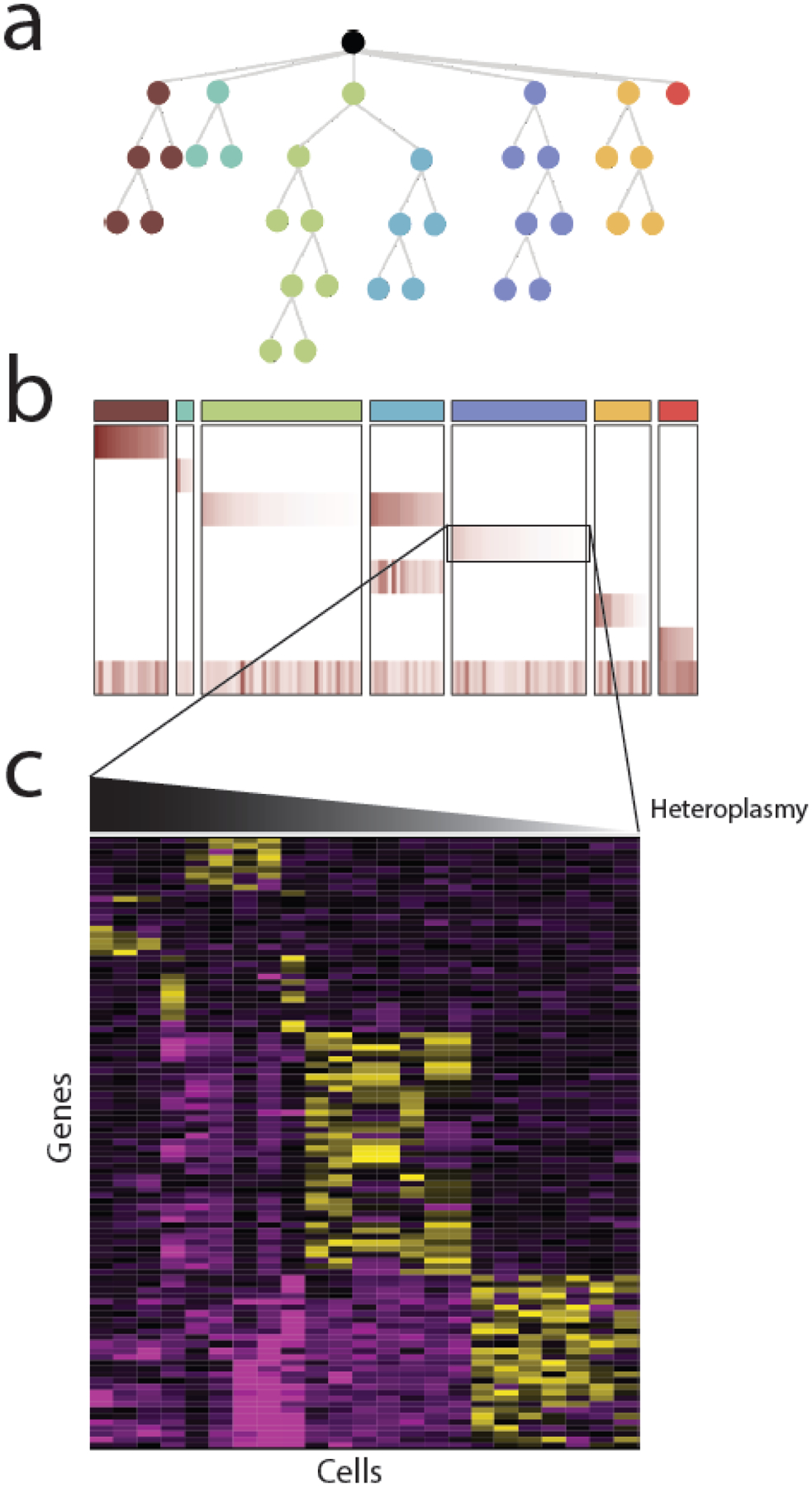
Most single cell analysis of mtDNA variants to date has been for the purposes of reconstructing evolutionary phylogenies, using mtDNA variants as cell-endogenous barcodes. (A) As cancer cells divide, they accumulate new somatic changes to nuclear and mitochondrial DNA that reflect their evolutionary phylogeny. (B) Evolutionarily related cells can be clustered into clones using information about the presence and heteroplasmy of heteroplasmic mtDNA variants. In htis example, a heatmap of the heteroplasmy levels of mtDNA variants reflects the evolutionary structure shown above in (A) The columns indicate each cell and rows indicate mtDNA variants. (C) Some mtDNA variants may not be silent, but rather may be associated with a particular phenotype. In this case, examination of nuclear gene expression patterns (e.g. derived from in single cell RNA-seq) may identify phenotypes associated with heteroplasmic dosage.
