Fig. 5.
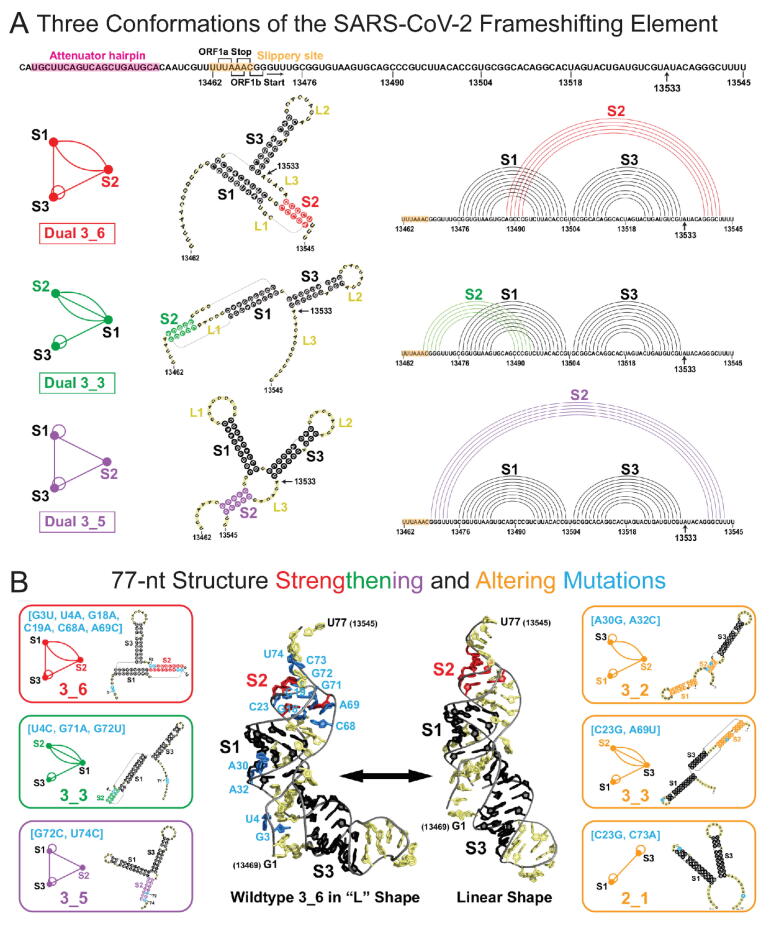
FSE sequence, three relevant 2D structures for the SARS-CoV-2 84 nt FSE (residues 13462–13545) emerging from the Schlick group’s work [84], [96] and mutants that transform the 3_6 motif and stabilize all conformers, using graph based models, 2D structure prediction, SHAPE structural probing, thermodynamic ensemble modeling, and molecular dynamics simulations [97]. (A) The −1 frameshifting moves the transcript UUU-UU*A(Leu)-AAC(Asn)-GGG at the second codon (asterisk) to start as AAA-CGG(Arg), so that translation resumes at CGG. Top: FSE sequence, with the attenuator hairpin region, the 7 nt slippery site, and A13533 labeled (C in SARS). The ORF1a end and ORF1b start codons for the overlapping regions are marked. For each 2D structure, 3_6 pseudoknot, 3_3 pseudoknot, and three-way junction 3_5 (unknotted RNA), we show corresponding dual graphs, 2D structures, and arc plots, with color-coded stems and loops. (B), Left: Mutants that stabilize each conformer; (B), Right: Double mutants that transform the 3_6 form into other RNAs. B, Middle: Straight to L-shape transition captured in the simulation for wild type 3_6 system, which is suppressed in the 3_6 stabilizing mutant.