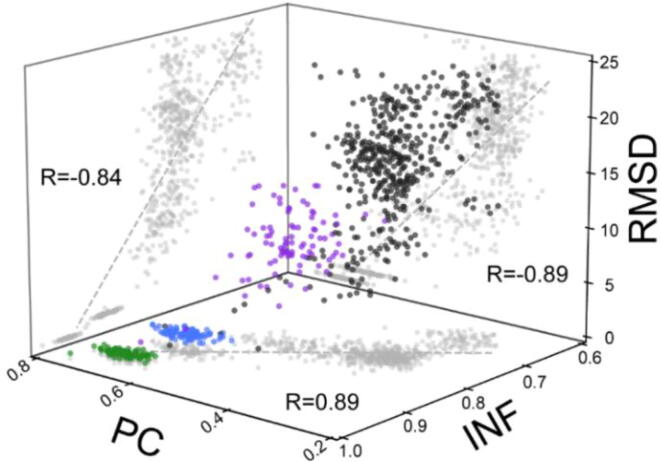
Fig. 8.
The figure shows the trends between the Pearson correlation, INF and RMSD for Lysine riboswitch (PDBID: 3DIG). The INF is a measure that captures native interactions, so the reference native structure has an INF of 1. The RMSD is the root mean squared deviation of the structure, so the reference native structure has an RMSD of 0. Structures in the green region are the decoys generated from a simulation of the native RNA with the backbone and base pairs restrained. The blue region represents a similar ensemble of structures, except that the base pairs are no longer restrained while the backbone atoms are restrained to maintain the global folding. Structures in the purple region are the decoys generated from coarse-grained simulations with the base pairs enforced, but no backbone restraints. The black region shows an ensemble of alternative (nonnative) low-energy 2D structure decoys generated from coarse-grained simulations. The projections of the decoy structures are shown as shadows in each 2D plane. We see that as the INF increases and RMSD decreases, i.e., as the structure approaches the native state, the Pearson correlation between the SHAPER prediction of the SHAPE reactivity and the experimental SHAPE reactivity of the structures increases, indicating that the model is successful in estimating the SHAPE reactivity for decoy structures. This may be used for predictive purposes. By sieving low-energy decoys, the model can improve predictions RNA structure prediction by removing structures that are incompatible with SHAPE data.