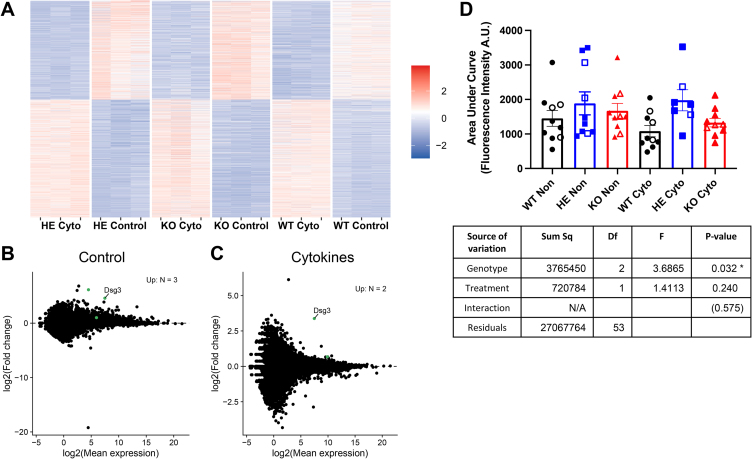
Figure 3.
Differentially regulated gene expression and increased calcium oscillations in islets treated with proinflammatory cytokines. (A–C) RNA-Seq analysis of 17 wks NOD.SCID.ZnT8 WT, HE and KO male islets; untreated and treated with proinflammatory cytokines for 24 h, N = 3 per group. Because WT and KO had similar phenotypes, these were averaged for differential expression analysis with DESeq2 (version 1.30.0). Differentially expressed genes were identified using the DESeq function with default parameters and genes for the comparison of the HE islets vs the average of the WT and KO islets were identified using the results function [results (contrast = list (HE_sample, c (WT_sample, KO_sample)), listValues = c (1, −1/2)]. (A) Heatmap of differentially expressed genes in individual replicates. (B, C) Differential expression analysis comparing averaged NOD.SCID.ZnT8 WT and KO with HE islets under control (B) and cytokine (C) conditions. Up-regulated genes with pAdj <0.05 are shown in green (n = 3 [control], n = 2 [cytokine]. (D) Islets (7–10 per group derived from 4 mice of each genotype (3 male and 1 female) were incubated for 24 h with or without proinflammatory cytokines, then loaded with a Fluo-4AM calcium indicator and imaged during stimulation with 11 mM glucose. The area under the curve for each islet is shown. Islets from male mice are shown as solid symbols and females as open symbols. 2-Way ANOVA was used to compare groups.∗p < 0.05.