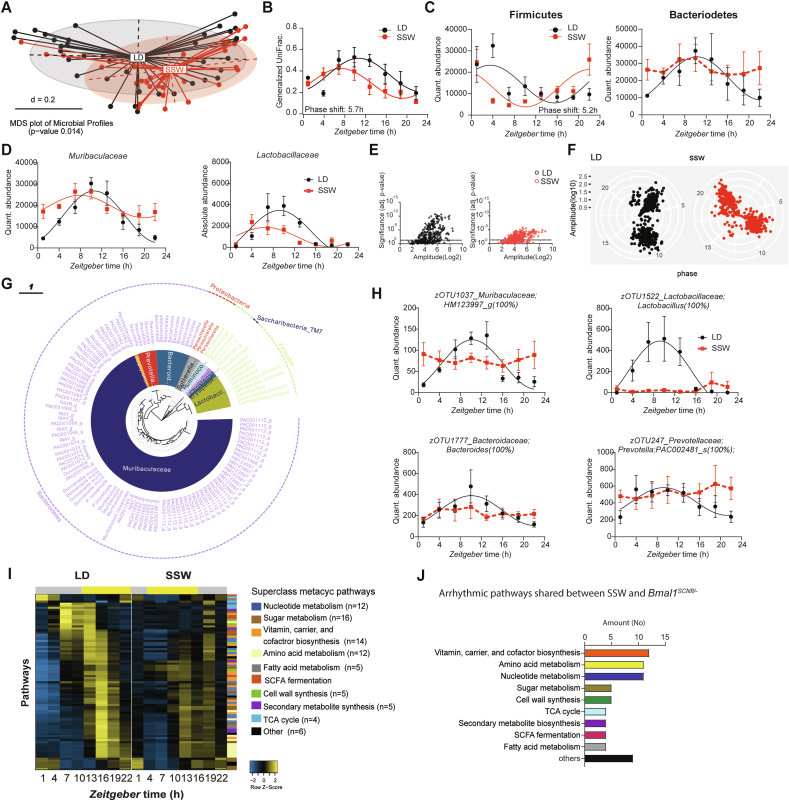
Figure 5.
Simulated shift work disrupts rhythmicity of microbiota composition and function (A) Beta-diversity MDS plot based on generalized UniFrac distances (GUniFrac) of fecal microbiota stratified by light condition. (B) Circadian profile of generalized unifrac distance normalized towards ZT1 of the controls. (C–D) Circadian profiles of the absolute abundance of major phyla (C) and families (D). (E) Significance and amplitude (based on JTK_CYCLE) of all zOTUs (E) and phase (based on cosine regression) distribution (F) in both genotype, dashed line represents adj. p-value = 0.05 (JTK_CYCLE). (G) Taxonomic tree of zOTUs losing rhythmicity in SSW based on quantitative analyses. Taxonomic ranks were indicated as phylum (outer dashed ring), then family (inner circle) and genera (middle names). Each zOTU is represented by individual branches. (H) Circadian profiles of absolute abundance of example zOTUs losing rhythmicity in SSW. (I) Heatmap representing MetaCyc Pathways predicted by PICRUST2.0 from zOTUs losing rhythmicity in SSW. Pathways are ordered by the phase of the control and normalized to the peak abundance of each pathway. We colored the pathways according to their sub-classes. (J) Bar chart representing the number of shared pathways losing rhythmicity in SSW and Bmal1SCNfl/- mice. Significant rhythms according to cosine-wave regression analysis (p-value ≤ 0.05) are visualized with a solid line, while data connected by dashed line indicate arrhythmicity. Significant phase shifts (p ≤ 0.05) are indicated with the number of hours of phase shift. n = 4–5 mice/time point/genotype. Data are represented as mean ± SEM.