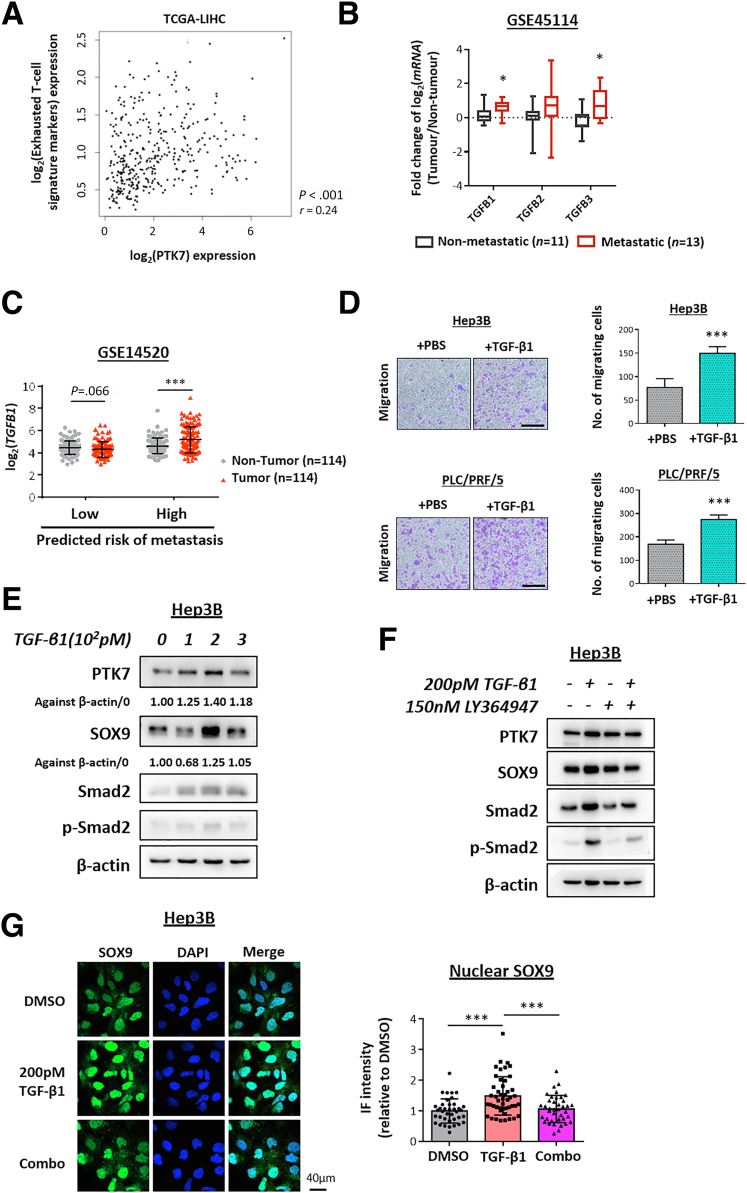
Figure 8.
In vitro analyses of TGF-β1–treated HCC cells. (A) PTK7 correlation with 6 exhausted T-cell signature markers (HAVCR2, TIGIT, LAG3, PDCD1, CXCL13, and LAYN) was examined by Pearson correlation using TCGA-LIHC data set and aided by the publicly available GEPIA2 analysis platform with all expression in terms of transcript per million. (B) Statistical comparison of the expression of TGFB1/2/3 in metastatic and nonmetastatic HCC by GSE45114 and the Student t test. (C) Same statistical evaluation of the GSE14520 data set as described in Figure 1B for the fold change in TGFB1 expression of paired tumor/nontumor HCC samples with different predicted risk of metastasis, by paired t test. (D) Representative images and the quantification of migrating cells by Transwell assay after TGF-β1 single treatment of 24 hours’ duration. Statistical comparisons by the Student t test. (E) Western blot validation of dosage-dependent up-regulation of PTK7 and SOX9 expression in Hep3B cells after a single treatment of recombinant TGF-β1 for a duration of 24 hours at various dosages. Quantification of PTK7 and SOX9 expression was performed by ImageJ. (F) Western blot validation of PTK7 and SOX9 expression in Hep3B cells after TGF-β1 single treatment or in combination with LY-364947, a TGF-βR1–specific inhibitor, for a duration of 24 hours. (G) Representative images of immunocytochemistry targeting SOX9 in Hep3B cells after TGF-β1 single treatment or in combination with LY-364947 (Combo) for a duration of 24 hours. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to locate the nuclei of stained cells. The average nuclear localization of the SOX9 was quantified by ImageJ and compared between treatment groups by the Student t test. DMSO, dimethyl sulfoxide. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).