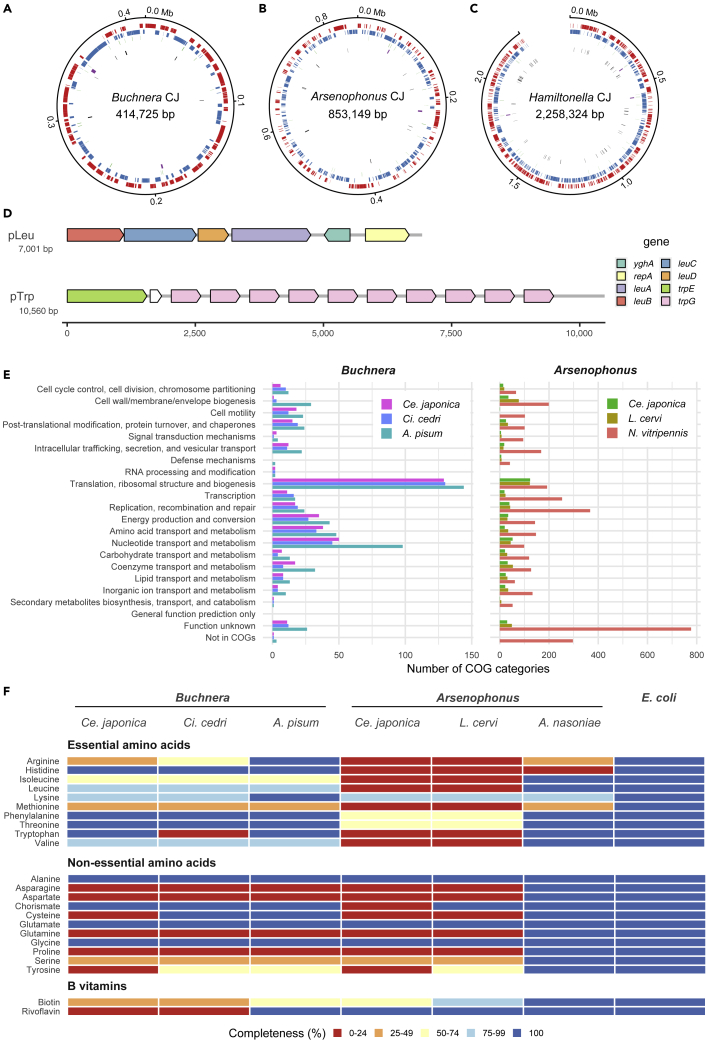
Figure 4.
Genomic features of Buchnera CJ and Arsenophonus CJ
(A) Circular Buchnera CJ genome.
(B) Circular Arsenophonus CJ genome.
(C) Linear Hamiltonella CJ genome. Outer to innermost rings correspond to (i) genome coordinates in kilobases; (ii) predicted protein-coding genes on the plus strand (red); (iii) predicted protein-coding genes on the minus strand (blue); (iv) transfer RNAs (green); (v) ribosomal RNAs (purple); (vi) pseudogenes (black) in (A–C).
(D) Gene orders of plasmids of Buchnera CJ pLeu and pTrp. Arrows indicate the direction of transcription. White arrows indicate pseudogenes.
(E) COG classification of protein-coding genes of Buchnera and Arsenophonus.
(F) Comparison of gene repertoires responsible for nutrient synthesis by Buchnera and Arsenophonus. E. coli is shown as an example of a free-living bacterium. Color blocks indicate the completeness of the minimal gene set for metabolic pathways: red, orange, yellow, light blue, and blue mean 0–24%, 25–49%, 50–74%, 75%–99%, and 100% of the completeness, respectively. In the aphid-Buchnera symbiosis, a metabolic collaboration is known for amino acid syntheses, where the host complements the steps missing from Buchnera as observed in the pathways of leucine, isoleucine, valine, and methionine syntheses (Shigenobu and Wilson, 2011; The International Aphid Genomics Consortium, 2010).