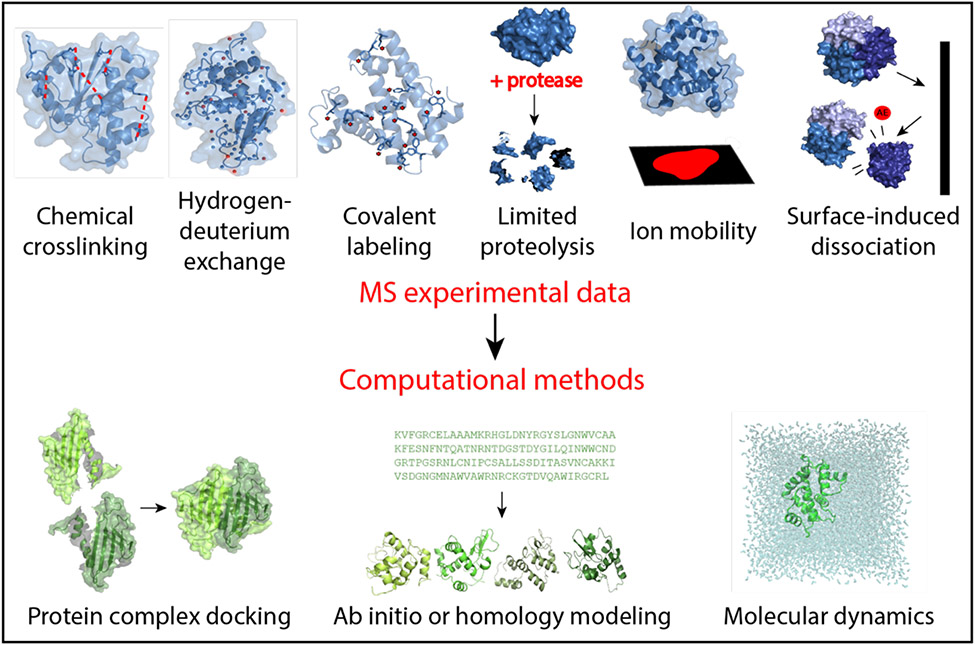
Figure 1.
Mass spectrometry-based methods and computational modeling explored in this review. Chemical crosslinking involves the modification of residues, commonly lysine, to provide information regarding spatial proximity. Hydrogen-deuterium exchange examines the exchange rate of amide hydrogens with deuterium solvent to give insight into solvent exposure and residue flexibility. Covalent labeling is reliant upon the irreversible covalent modification of residues, illuminating solvent exposure and topology. Limited proteolysis uses a protease enzyme to cleave proteins into fragments based on solvent exposure. Ion mobility is used to investigate shape and size of proteins based on the collision cross sectional area. Appearance energies (AE) can be deduced from surface-induced dissociation, which is used to study the stoichiometry and connectivity of protein complexes. Data from these techniques is then incorporated into computational modeling techniques such as protein-protein docking to examine complexes, structure prediction via ab initio or homology modeling, and molecular dynamics based on experimental restraints.