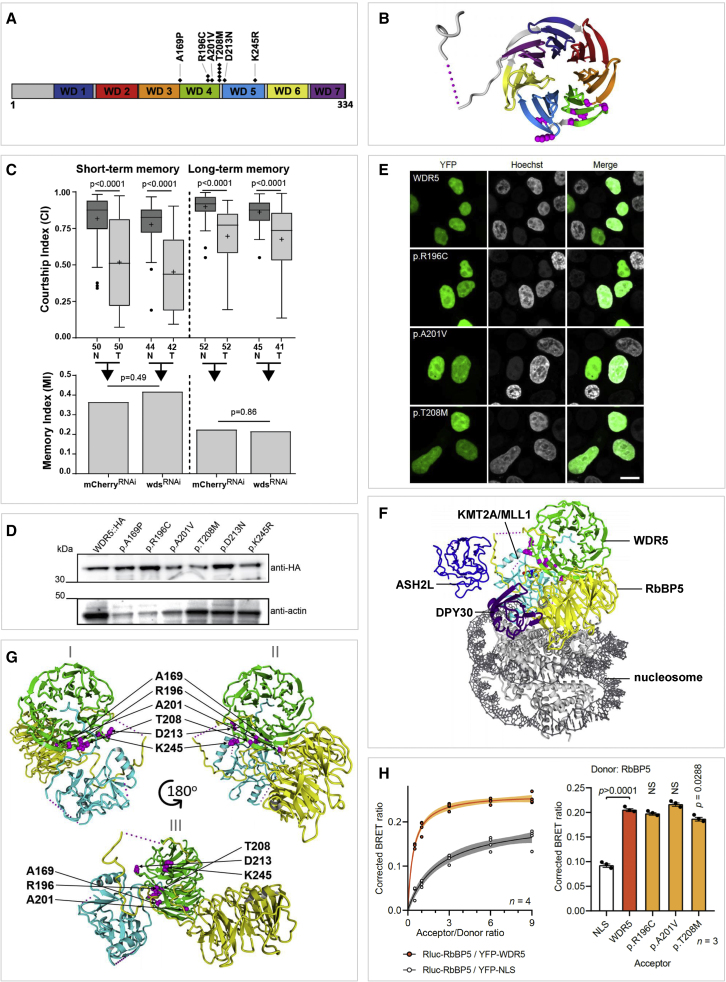
Figure 3.
In silico, in vitro, and in vivo studies of the effect of WDR5 missense variants
(A) Linear structure of WDR5 protein (334 amino acids) with the seven different WD40 domains and all identified missense variants shown; in total, six different missense variants were found in 11 individuals: one variant (p.(Thr208Met)) was found in five unrelated individuals and another variant (p.(Arg196Cys)) in two individuals.
(B) Three-dimensional visualization of WDR5 (PDB: 2GNQ); locations of the amino acids involved in missense variants are shown with magenta balls. Colors of the different domains match with the colors used in (A).
(C) Courtship memory was assessed in mushroom-body-specific wds RNAi knockdown flies (wdsRNAi) compared with controls expressing an RNAi against mCherry (mCherryRNAi). Boxplots show the distribution of courtship indices (CIs) for naive (N) and trained (T) flies (top panel). Memory was observed when a significant reduction in CI occurred between naive and trained conditions (Mann-Whitney test). + indicates the mean. N is indicated along the x axis. Bar graphs show the memory indices (MIs), which are single values derived from the above CIs (indicated by arrows) according to the formula: MI = ( CInaive − CItrained)/ CInaive. MIs were consistent between controls and wdsRNAi lines (randomization test, 10,000 bootstrap replicates).
(D) Detection of WDR5 reference and variant proteins in adult flies by western blot. Representative bands for HA-tagged WDR5 reference and variant proteins at 36.6 kDa (top) along with the actin loading control 41 kDa (bottom). UAS-WDR5::HA reference and variant transgenes were expressed ubiquitously using Actin-Gal4.
(E) Direct fluorescence micrographs of nuclei of HEK293T/17 cells expressing YFP-WDR5 fusion proteins (green). Nuclei were stained with Hoechst 33342 (blue). Scale bar: 10 μm.
(F) WDR5 (green) is shown as part of the core COMPASS complex, with RbBP5 (yellow), ASH2L (blue), DPY30 (purple), and KMT2A (cyan) (PDB: 6KIV). The nucleosome is shown in gray. The locations of affected amino acids in individuals with missense variants are shown with magenta balls.
(G) WDR5 (green; p.33–332) is shown together with RbBP5 (yellow; p.1–380) and KMT2A (cyan; p.3764–3969) as part of the core COMPASS complex (PDB: 6KIV). The locations of affected amino acids in individuals with missense variants are shown with magenta balls from three different angles facing the WIN site (I), the WBM site (II), and a side between WIN and WBM (III).
(H) BRET assays for WDR5-RbBP5 interaction in live cells. Left, mean BRET saturation curves ±95% confidence interval fitted using a nonlinear regression equation assuming a single binding site (n = 4; y = BRETmax ∗ x/(BRET50/x); GraphPad) showing a strong BRET signal for Rluc-RbBP5 with YFP-WDR5. Right, corrected BRET values measured with an acceptor/donor ratio of 1:1 (n = 3, one-way ANOVA and post-hoc Bonferroni test). NLS, YFP fused to a C-terminal nuclear localization signal as control protein. p values show significance for the comparison of WDR5 variant with the WDR5 wild-type.