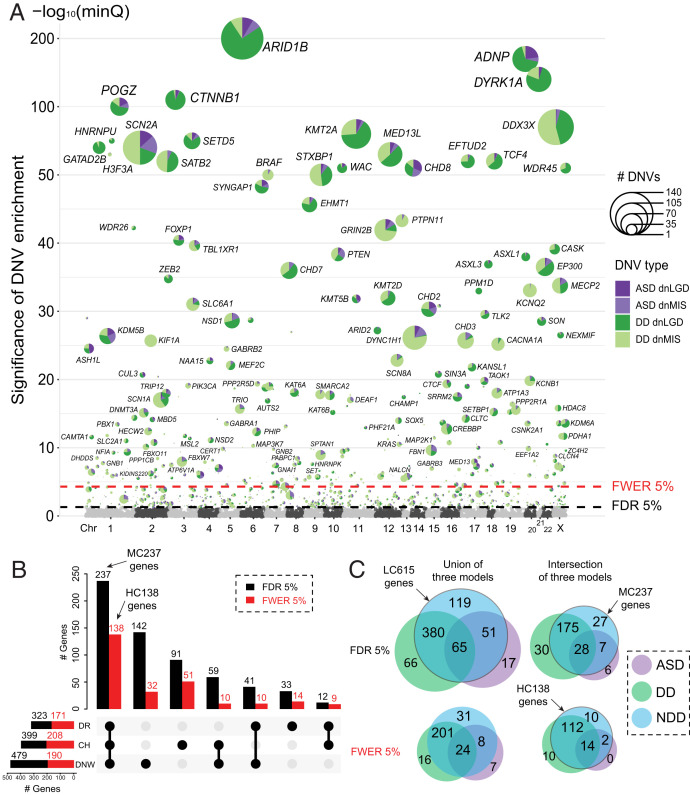
Fig. 2.
De novo enrichment analysis and significant genes by model and phenotype. (A) The smallest q value (minQ) after Benjamini-Hochberg correction of each gene across the three models was plotted in alphabetic order of gene name by chromosome. The LC615 genes reaching union FDR 5% significance were plotted with the number of dnLGD and dnMIS variants in the ASD and DD cohorts scaled in pie charts; the HC138 genes reaching the intersection FWER 5% significance were additionally labeled with gene name. (B) The number of genes reaching FDR 5% (black bar) and FWER 5% (red bar) significance identified by each of the three models (DR: denovolyzeR; CH: CH model; DNW: DeNovoWEST) in the combined NDD set. (C) Cohort overlap among low-confidence (n = 615, FDR 5%) and high-confidence (n = 138 FWER 5%) gene sets considering the ASD and DD cohorts separately and as one group (NDD). Genes were compared based on the union of three models for DNV enrichment versus only those that were observed by all three (intersection).