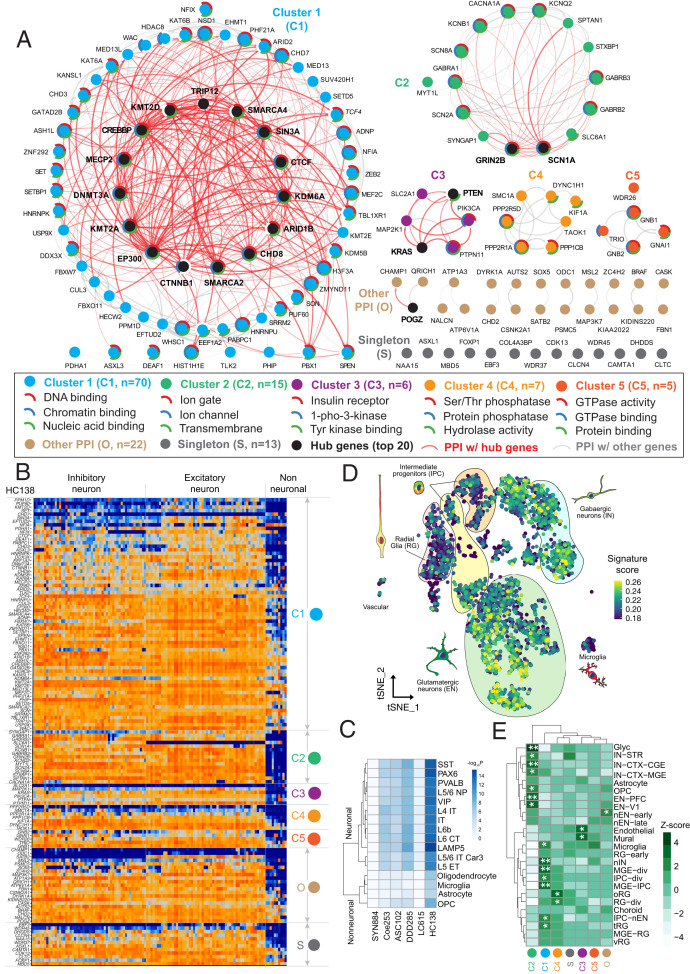
Fig. 6.
PPI analysis and panneuronal expression of the highest confidence genes. (A) PPI analysis identified five main clusters (C1 to C5), as well as 22 genes in another smaller PPI group (O) and 13 singleton genes (S) using STRING for the HC138 genes with the highest confidence. The top three GO functions were indicated by the pie chart in color (top 1 in red, top 2 in blue, and top 3 in green), if applicable, outside each gene dot with a short name in the legend. We also defined 20 top hub genes (black dot and bolded name) supported by at least half of 12 statistical methods in cytoHubba (37). Red arcs designate PPI with hub genes; gray arcs indicate PPI only between non-hub genes. The degree of the color and the width of the arc indicate the degree of the interaction. (B) Expression heatmap of the HC138 genes in 120 cell types identified across 6 human neocortical areas grouped by PPI clusters with higher expression (orange) and lower expression (blue). The cell types were grouped by transcriptomic similarity, and the major branches correspond to inhibitory and excitatory neurons and nonneuronal cells. Labels on the Right Side indicate clusters based on PPI analysis, and gene names are on the Left Side. (C) Heatmap of −log10-transformed P values (Bonferroni corrected for multiple testing) of a Kolmogorov-Smirnov test for the difference in the expression levels of each gene set (rows) in each cell subtype (columns, full name described in Materials and Methods) compared to a control set of genes (HC138 and LC615 are the gene sets with the HC and LC identified in this study; DDD285 (18), ASC102 (17), and Coe253 (19) are significant genes reported previously; SYN884, the control set, includes 844 genes with dnSYN variants [n > 2] in the 46,612 NDD samples in this study). (D) Gene signature score of the HC138 genes computed per cell. IN: interneurons; EN: excitatory neurons; IPC: intermediate progenitor cells; MGE: medial ganglionic eminence; CGE: caudal ganglionic eminence; OPC: oligodendrocyte progenitor cells; tRG: truncated radial glia; oRG: outer radial glia; vRG: ventral radial glia; CTX: cortex; V1: visual cortex; PFC: prefrontal cortex; STR: striatum. (E) The heatmap shows the SD from the mean expression value of each cluster of genes. Positive values are up-regulated compared to the mean, and negative values are down-regulated compared to the mean. Significance is derived from bootstrapping and labeled with asterisk (*P < 0.05, **FDR P < 0.05 after Benjamini-Hochberg correction). The HC138 genes were used as the background gene set.