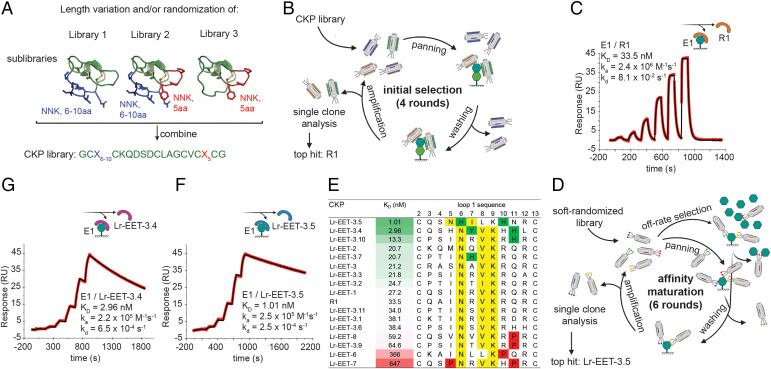
Fig. 1.
CKP library design and phage selection strategy for discovery of high affinity binders to LRP6 E1. (A) Design of CKP library. The randomized regions are highlighted in blue and red for loop 1 and 5, respectively, on the native EETI structure (Protein Data Bank [PDB]: 1W7Z). NNK: randomization by NNK-codon incorporation; aa: amino acid. (B) Schematic of phage display workflow during initial selections. (C, F, and G) Affinity determination of CKP hits by SPR (measured by single-cycle kinetics). E1 was immobilized on sensor chips, and different CKP concentrations were injected. Sensograms of the the parental CKP R1 (C), and the two affinity-matured CKPs Lr-EET-3.5 (F) and Lr-EET-3.4 (G). Fit to a 1:1 model is shown in black. (D) Schematic workflow of phage display during affinity maturation. (E) Comparison of affinity and sequence of affinity matured CKPs. CKPs identified by the affinity maturation and the parental CKP R1 are sorted by affinity (green-white-red gradient), with their loop 1 sequences shown. Nx(I/V)K motifs are highlighted in yellow, and residues detrimental or beneficial to binding are highlighted in red and green, respectively.