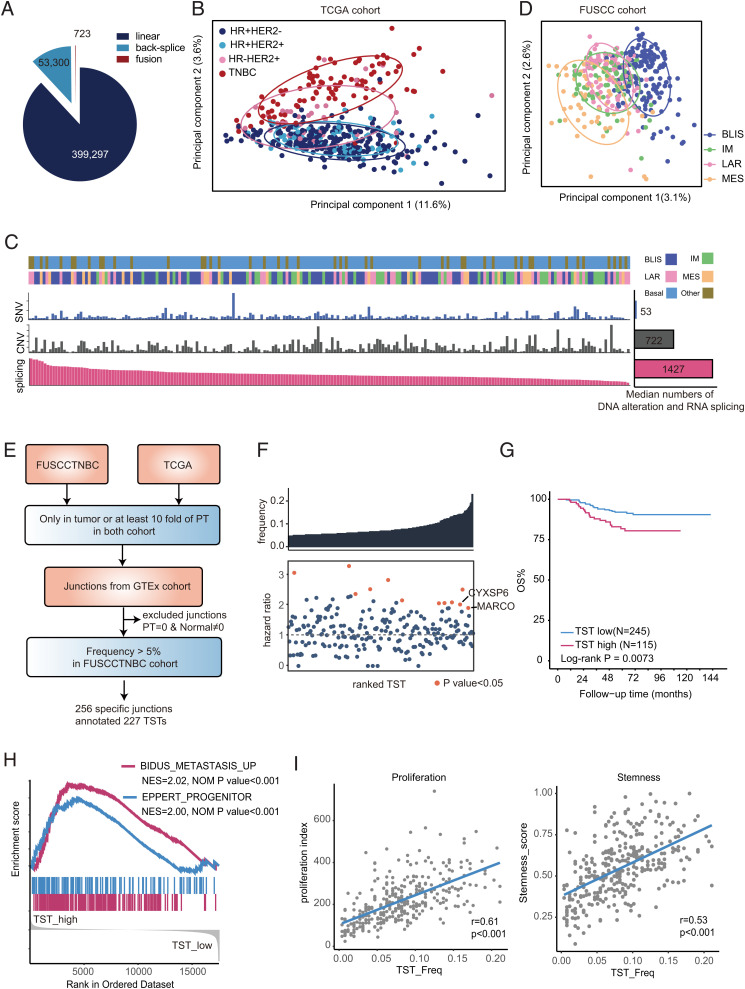
Fig. 1.
The landscape of RNA splicing in TNBC. (A) The fraction and number of linear, fusion, and back-splice junctions in the FUSCCTNBC cohort. (B) PCA of splicing junctions of breast cancer tissues in the TCGA cohort (n = 548) using the CPT of splicing junctions. Ellipses are drawn for the 68% confidence zone. HR, hormone receptor. (C) The number of DNA or RNA alterations across TNBCs in the FUSCCTNBC cohort. The samples were ordered by the number of alternative splicing events per sample from high to low. The bar plot shows the median number of DNA or RNA alterations. SNV, single nucleotide variants; CNV, copy number variation. (D) PCA of splicing junctions of TNBC in the FUSCCTNBC cohort (n = 360) using the CPT of splicing junctions. Ellipses are drawn for the 68% confidence zone. (E) The workflow for the identification and validation of TSTs in TNBC. (F) Detection frequency and hazard ratio of each TST. The Top Box indicates the detection frequency. The Bottom Box shows the hazard ratio of DMFS, and orange points represent survival-associated TSTs. The P values were calculated using the Cox regression model. (G) Kaplan-Meier plots for the overall survival (OS) of patients with high TST burden (>22 transcripts) and low TST burden (≤22 transcripts). The P values were calculated using the log-rank test. (H) GSEA plots showing the enrichment of metastasis- and progenitor-related gene sets in high-TST-burden patients compared to low patients. (I) Correlation of TST burden with proliferation and stemness scores in the FUSCCTNBC cohort. Pearson’s correlation coefficient was used to determine the correlation (SI Appendix, Figs. S1 and S2 and Datasets S1 and S2).