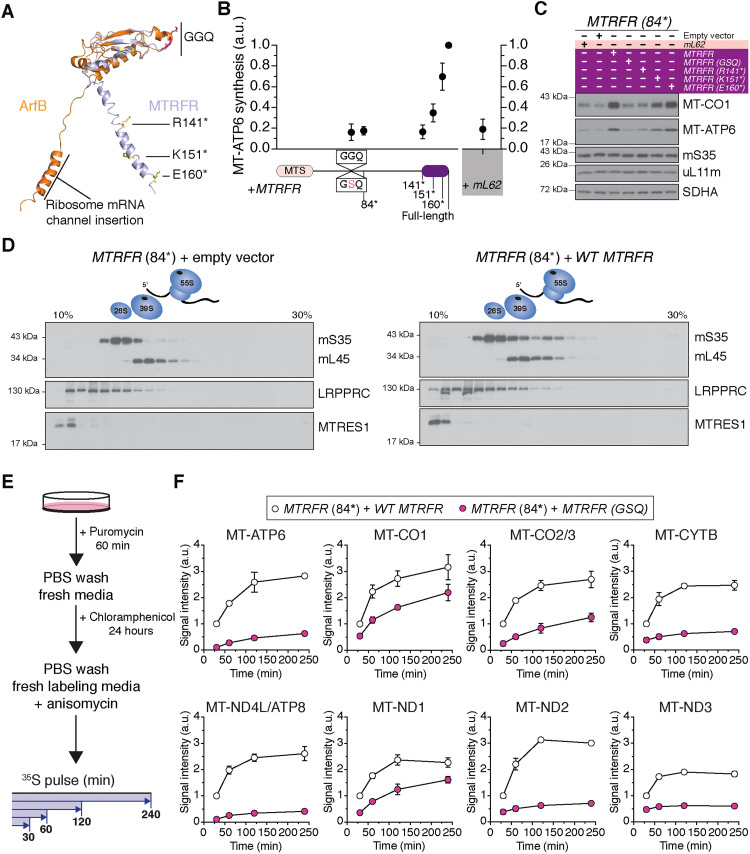
Fig. 8. Functional characterization of MTRFR in mitochondrial protein synthesis.
(A) Structural overlay of MTRFR (C12orf65) [Protein Data Bank (PDB) 7A5H] (35) with ArfB (PDB 7JSS) (32). The catalytic GGQ motif of the release factors and the position of truncation mutations in MTRFR are indicated. (B) 35S-metabolic labeling of MT-ATP6 synthesis in MTRFR-deficient (84*) fibroblasts stably transduced with cDNAs of the indicated MTRFR alleles and wild-type mL62 (ICT1). Data represent the means ± SD from four independent experiments. MTS, mitochondrial targeting sequence; GGQ, glycine-glycine-glutamine catalytic domain of the release factor. (C) Representative immunoblotting of whole cell lysates from (B). (D) Immunoblotting of fractions isolated from sucrose density gradient separation of mitochondrial ribosomes from human patient fibroblasts carrying biallelic frameshift variants that terminates MTRFR at codon 84, rendering the protein functionally null. Cells were stably transduced with an empty retroviral vector or a wild-type cDNA of MTRFR. The data are representative of multiple independent experiments. (E) Schematic of workflow for metabolic labeling of mitochondrial protein synthesis with 35S-methionine/cysteine in human patient fibroblasts from (B). (F) Quantification of mitochondrial protein synthesis over an extended pulse labeling period. Data represent the means ± SD from three independent experiments.