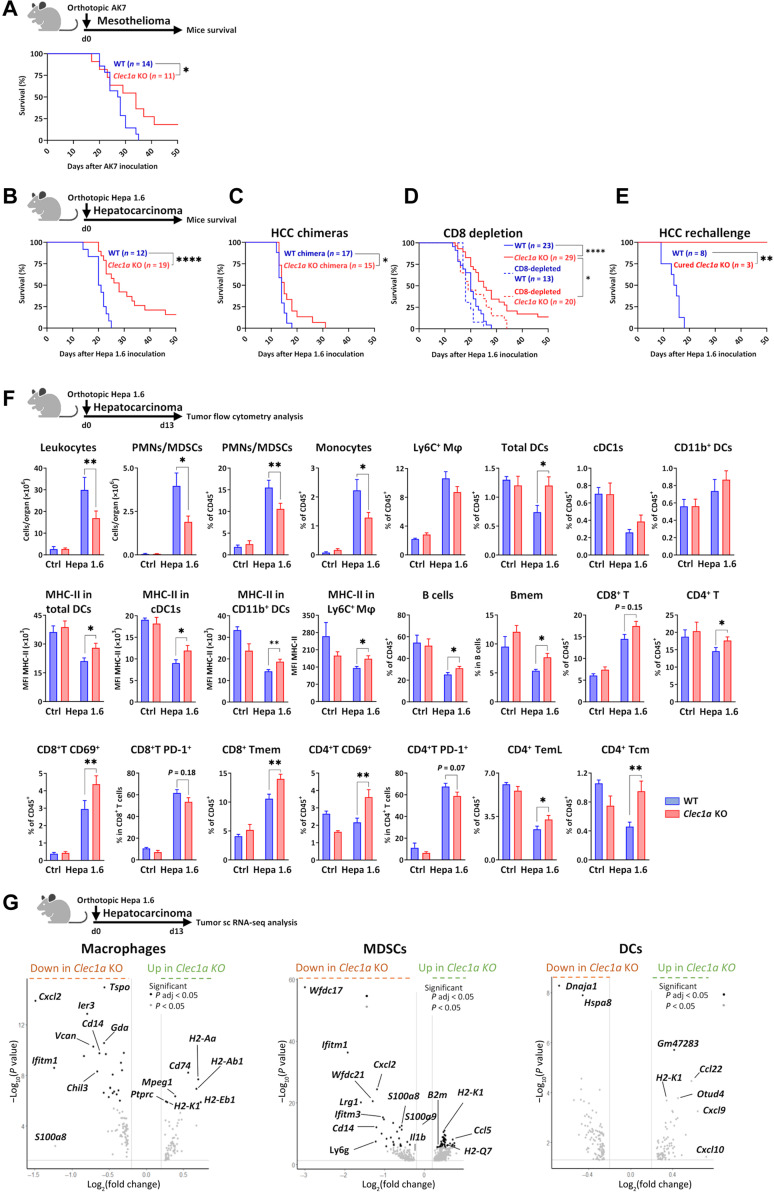
Fig. 4. Absence of Clec1a enhances antitumor immunity in mouse orthotopic models.
(A) Survival curve of WT and Clec1a KO mice with pleural mesothelioma (AK7) (n = 11 to 14, log-rank test, *P < 0.05). (B) Survival curve of WT and Clec1a KO mice with Hepa 1.6 orthotopic HCC (n = 12 to 19 of three independent experiments, log-rank test, ****P < 0.0001). (C) Survival curve of chimeric mice with the absence of CLEC-1 only in the hematopoietic compartment (Clec1a KO chimeras) or WT chimeras with HCC (n = 15 to 17 of two independent experiments, log-rank test, *P < 0.05). (D) Survival curve of WT, Clec1a KO, CD8-depleted WT, and CD8-depleted Clec1a KO mice injected with HCC (n = 13 to 29 of two independent experiments, log-rank test, *P < 0.05 and ****P < 0.0001). (E) Survival curve of HCC-cured Clec1a KO rechallenged in the spleen with HCC at day 90 after first inoculation (WT naïve mice were used as controls) (n = 3 to 8, log-rank test, **P < 0.01). (F) Total number and/or percentage of leukocytes, PMNs/MDSCs (CD11b+Ly6G+MHC-II−), monocytes (CD11b+Ly6C+MHC-II−), macrophages (Mφ) (CD11b+F4/80+Ly6C+), total DCs (CD11c+MHC-II+), cDC1 (CD11c+MHC-II+CD11b−XCR1+), DCs CD11b+ (CD11c+MHC-II+CD11b+), B memory (Bmem) (CD19+CD45R+CD24+CD38−), CD8+ or CD4+ T (CD3+CD4/8+), CD69+, PD-1+, T memory (Tmem) (CD44+), T effector-memory Late (TemL) (CD44+CD62L−CD27−), and T central memory (Tcm) (CD44+CD62L+) CD8+ or CD4+ T cells evaluated by flow cytometry in the tumor-burdened livers of WT and Clec1a KO mice at day 13 after HCC [data are expressed in cell count, in the percentage of CD45+, in the percentage in particular cell subset or in MFI of MHC-II expression] (n = 5 for controls and n = 15 for Hepa 1.6–treated mice, means ± SEM of three independent experiments, unpaired t test, *P < 0.05 and **P < 0.01). (G) Volcano plot representations of differential gene expression analysis by single-cell RNA sequencing (scRNA-seq) of macrophage, MDSC, and DC clusters from WT and Clec1a KO mice at day 13 after HCC. Data represent the −log10 (P values) versus the log2 (fold change), and genes are colored on the basis of their significance.