Figure 5.
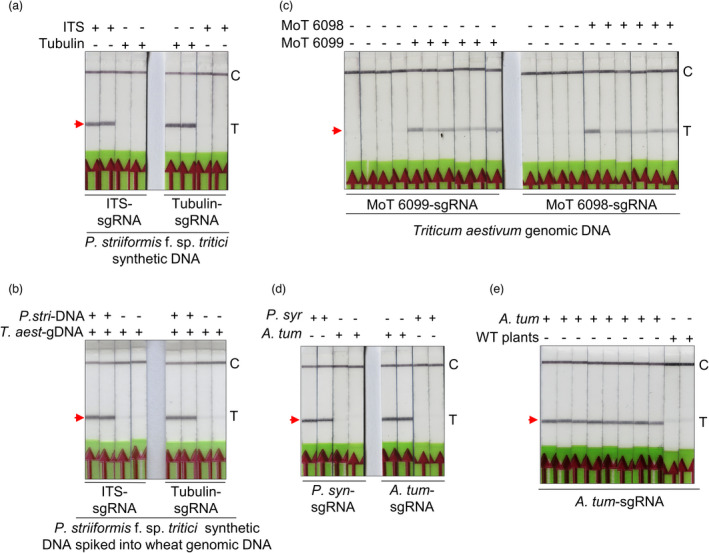
Bio‐SCAN efficiently detects pathogenic fungi of wheat and bacterial pathogens in Nicotiana benthamiana. (a) Bio‐SCAN detects specific sequences (ITS and β‐tubulin) of Puccinia striiformis f. sp. tritici in synthetic DNA. The pathogen‐specific sequence was detected on LFA strips via RPA‐based Bio‐SCAN assay. A nonspecific sgRNA was used as a control. (b) BioSCAN‐based detection of Puccinia striiformis f. sp. tritici in wheat genomic DNA. Synthetic Puccinia striiformis f. sp. tritici DNA was spiked into wheat genomic DNA and detected via RPA‐Bio‐SCAN. P. stri (Puccinia striiformis f. sp. tritici): T. aest (Triticum aestivum). (c) Detection of Magnaporthe oryzae Triticum in wheat samples. Wheat tissues infected with M. oryzae triticum were used for total genomic DNA isolation. The extracted genomic DNA was subjected to an RPA‐based Bio‐SCAN assay. Bio‐SCAN specifically detected both conserved sequences (MoT 6099 and MoT 6098) of M. oryzae Triticum in samples isolated from infected wheat plants. Noninfected wheat tissues were used as a control. (d) Bio‐SCAN detects specific sequences, AvrE of Pseudomonas syringae and ChvA of Agrobacterium tumefaciens in synthetic DNA. P. syn (Pseudomonas syringae); A. tum (Agrobacterium tumefaciens). The pathogen‐specific sequence was detected on LFA strips via RPA‐based Bio‐SCAN assay. A nonspecific sgRNA was used as a control. (e) BioSCAN‐based detection of Agrobacterium tumefaciens in the genomic DNA of N. benthamiana plants. Total DNA is isolated from agroinfiltrated N. benthamiana plants and detected via RPA‐Bio‐SCAN. Noninfiltrated N. benthamiana tissues were used as a control. C, control line; T, test line. The arrowheads indicate the location of the expected band at the T line on the LFA strip. [Correction added on 27 October 2022, after first online publication: Figure 5 has been corrected in this version.]
