Figure 2.
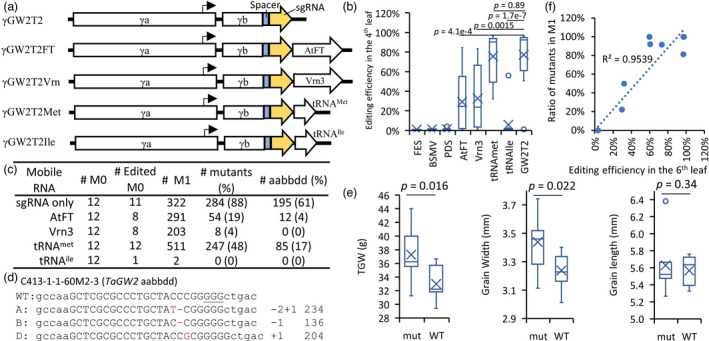
The effects of mobile RNA fusion on gene editing efficiency induced by the BSMV‐sgRNA constructs. (a) The structure of the BSMV‐GW2T2‐based constructs targeting the TaGW2 gene. The GW2T2‐sgRNA was fused with the AtFT (γGW2T2FT), Vrn3 (γGW2T2Vrn), tRNAmet (γGW2T2Met), and tRNAile (γGW2T2Ile). The open reading frames (ORFs), promoters, and untranslated regions are shown as open boxes, arrows, and hatched boxes, respectively. Orientation of ORFs is depicted by arrow‐shaped boxes. (b) Somatic editing efficiency induced by the BSMV‐ sgRNA constructs with and without mobile RNA in the noninoculated 4th leaf of the C413 line. The mutation frequency at the GW2T2 target was calculated by combining data from all three genomes. The significance of differences between the respective gRNAs with and without mobile RNAs was tested using the Student's t‐test (P‐values are shown above the plot). (c) The summary of BSMV‐sgRNA inoculation experiment showing the number of inoculated plants and the frequency of mutations in TaGW2. The somatic mutations in the M0 generation were evaluated using DNA extracted from the noninoculated 4th leaf. The number of mutants in the M1 generation corresponds to the number of plants carrying mutations in at least one homoeolog of TaGW2. The genotypes of triple TaGW2 mutants with mutations in each of the homoeologous wheat genomes A, B, and D are shown as lower‐case letters aabbdd. (d) The alignments of representative NGS reads from M1 plant C413‐1‐1‐60 M2‐3, which carries mutations in each of the homoeologous copies of TaGW2 from three wheat genomes. The level of divergence between the wheat genomes allows for homoeolog‐specific alignment of NGS reads. The deleted and inserted nucleotides are shown in red. The number of deleted (−1 or −2) or inserted (+1) nucleotides are shown on the right of each read type. The number of wild‐type (WT) and mutated reads aligned to each genome is shown on the right side of mutation types. The PAM sequence is underlined. (e) Phenotypic effects of BSMV‐sgRNA induced mutations in TaGW2. The thousand grain weight (TGW), grain width, grain length of triple mutants (aabbdd, n = 7), and wild‐type plants (AABBDD, n = 7) are compared. The Student's t‐test P‐values are shown above the plot. (f) Relationship between the proportion of TaGW2 mutants in the M1 generation and somatic editing efficiency evaluated in the 6th leaf at the booting stage of M0 plants. Each data point stands for an individual plant.
