Figure 3.
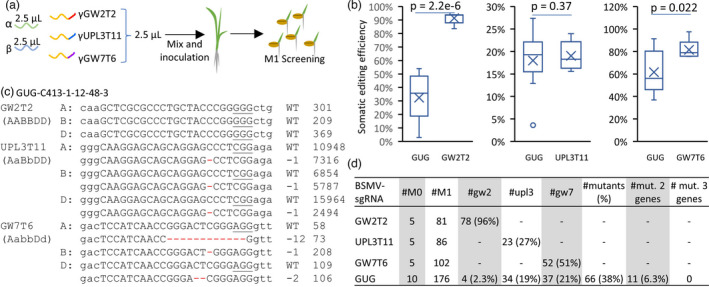
Multiplex gene editing by inoculating with the pooled BSMV‐sgRNAs. (a) A schematic pipeline of the BSMV‐based multiplex editing using the sgRNA pool targeting TaGW2 (GW2T2‐sgRNA), TaUPL3 (UPL3T11‐sgRNA), and TaGW7 (GW7T6‐sgRNA). (b) Comparison of somatic editing efficiency at each target site in plants inoculated with the BSMV‐GUG pool or individual BSMV‐sgRNAs. The data from all three genomes were combined to calculate the somatic editing efficiency. The multiplex and single target editing efficiencies were compared using the Student's t‐test (P‐values are shown above the plots); NS – P > 0.05, * – P < 0.05, *** – P < 0.0001. (c) Alignment of representative NGS reads from the M1 plants derived from the BSMV‐GUG inoculated plants. The NGS reads were aligned to the A, B, and D homoeologs of the TaGW2, TaUPL3, and TaGW7T6 genes. The deleted nucleotides are shown in red. The number of deleted nucleotides is shown on the right of the reads. The number of wild‐type (WT) and mutated reads is shown on the right side of mutation types. The PAM sequences are underlined. (d) The number of plants carrying mutations in one, two, or all three genes obtained by inoculating with BSMV‐GUG and the individual BSMV‐sgRNAs.
