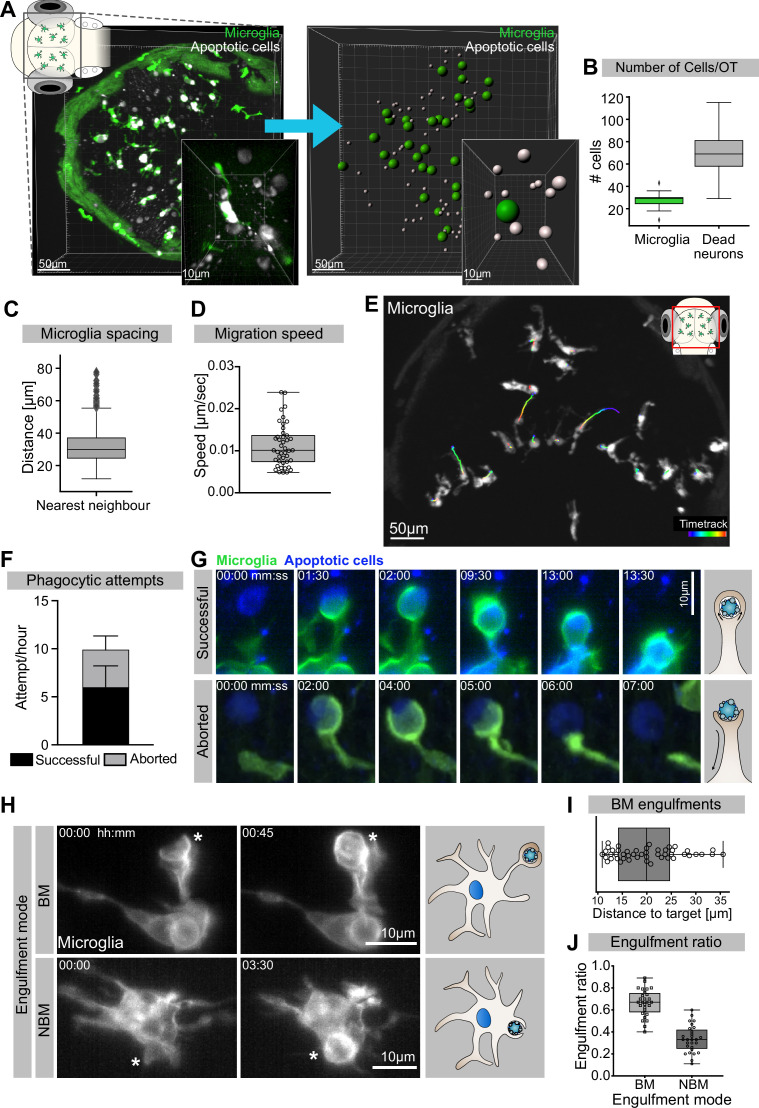
Figure 1. Mechanisms of neuronal efferocytosis by microglia.
(A) Left: representative image of the OT of a 3.5-day post-fertilization (dpf) embryo with microglia (green; Tg(mpeg1:eGFP-caax)) and dying neurons (grey; Tg(nbt:dLexPR-LexOP:secA5-BFP)). Right: automated spot detection of left image to determine the spatial position of microglia and apoptotic cells. (B) Number of microglia and dead neurons within the OT (N = 31). (C) Distance from each microglia to its closest neighbour (N = 31). (D) Microglia migration speed, measured by tracking microglia for 2 hr (N = 2, 20–23 microglia analysed per zebrafish). (E) A representative image of a 3-dpf zebrafish brain, showing microglia (grey; Tg(mpeg1:eGFP-caax)) and their trajectory/track over 1 hr. (F) The total number of phagocytic attempts initiated by microglia per hour; stacked barplot shows the proportion of successful and aborted phagocytic attempts (N = 4, 5–9 microglia analysed per fish). (G) Upper panel: microglia (green; Tg(mpeg1:eGFP-caax)) phagocytic cup that results in the successful formation of a phagosome around a dead neuron (blue; Tg(nbt:dLexPR-LexOP:secA5-BFP)). Lower panel: phagocytic cup formation where the phagocytic attempt is aborted. (H) Microglia (grey; Tg(mpeg1:eGFP-caax)) phagocytosis happens at two locations; upper panel: phagosome forms at the tip of a long cellular extension. Lower panel: phagosome forms directly at the cell soma. Full time lapse is found in Figure 1—video 2. (I) Length of successful phagocytic branches during branch-mediated (BM) engulfments (N = 3, n = 7, 5–16) engulfments analysed per microglia. (J) Ratio between BM and non-branch-mediated (NBM) engulfments (N = 4, 5–9 microglia analysed per fish). Bars represent mean +/- SD (F). Boxplots depict mean and 1.5x interquartile range (B, C) or mean +/- min to max values (D, F, I, J). N refers to the number of zebrafish and n to the number of microglia examined.