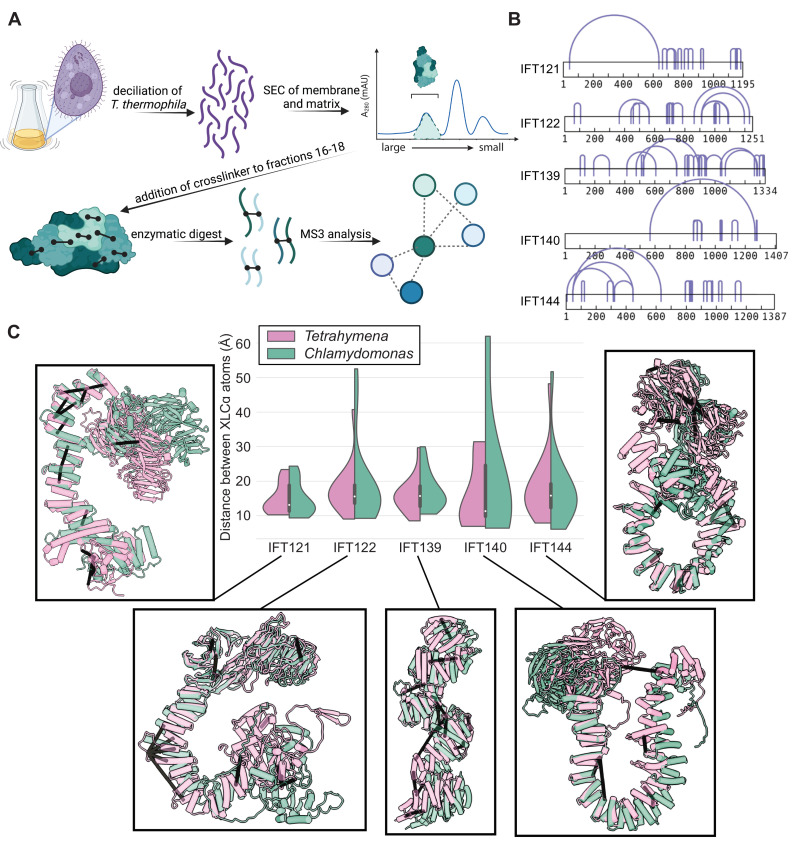
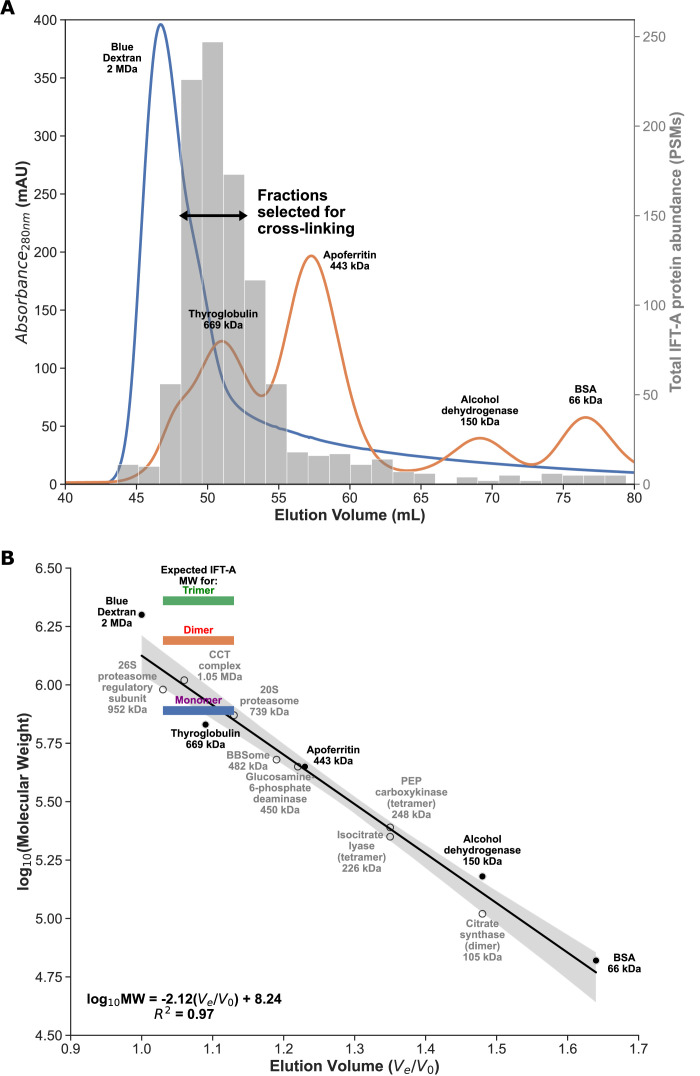
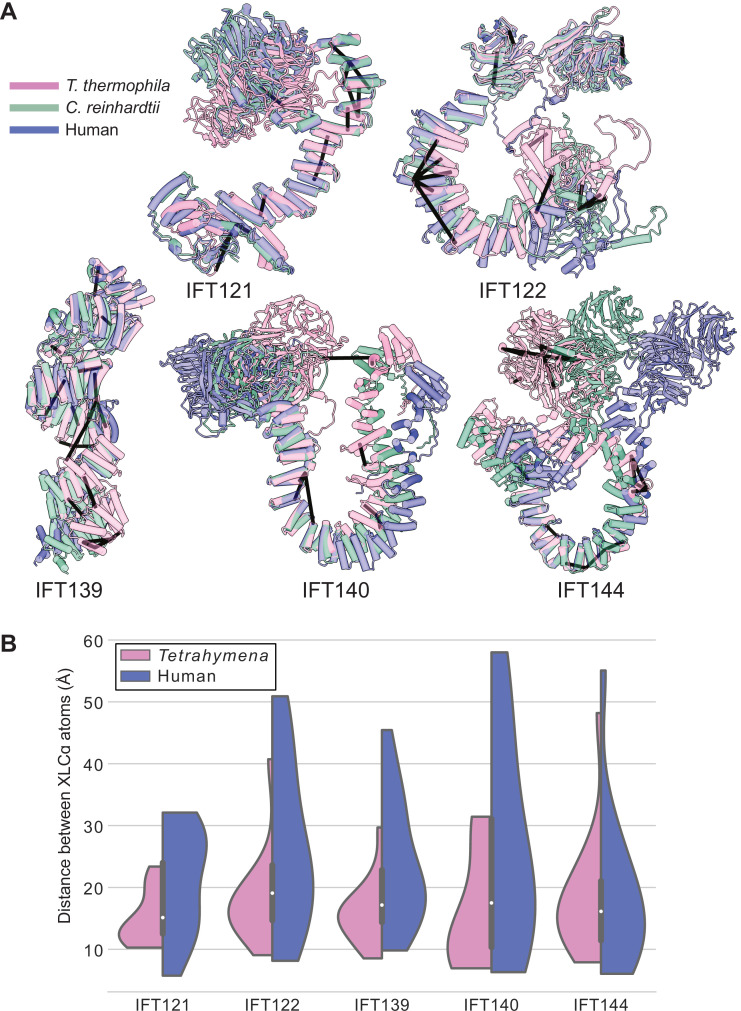
Figure 1. AlphaFold2 structures of the IFT-A subunits are supported by experimentally-determined intramolecular cross-links.
(A) Sample preparation protocol for obtaining an enriched sample of endogenous IFT-A from T. thermophila. The sample preparation was followed by chemical cross-linking using DSSO and tandem (MS2/MS3) mass spectrometry to identify cross-linked peptides. Image created with BioRender.com. (B) Bar diagrams highlight the extensive intramolecular DSSO cross-links (purple arcs connecting cross-linked amino acid pairs) within each of the IFT-A subunits (bars, numbers denote amino acid positions). (C) Violin plots of the distance between Cɑ atoms of chemically cross-linked residues. Surrounding images show locations of intramolecular crosslinks (black bars) on aligned AlphaFold2 predicted models of IFT-A subunits from Chlamydomonas reinhardtii (green) and Tetrahymena thermophila (pink). A maximum distance of 35 Å between Cɑ atoms is expected for DSSO cross-links. 97% of intramolecular cross-links are satisfied for T. thermophila and 94% for C. reinhardtii.