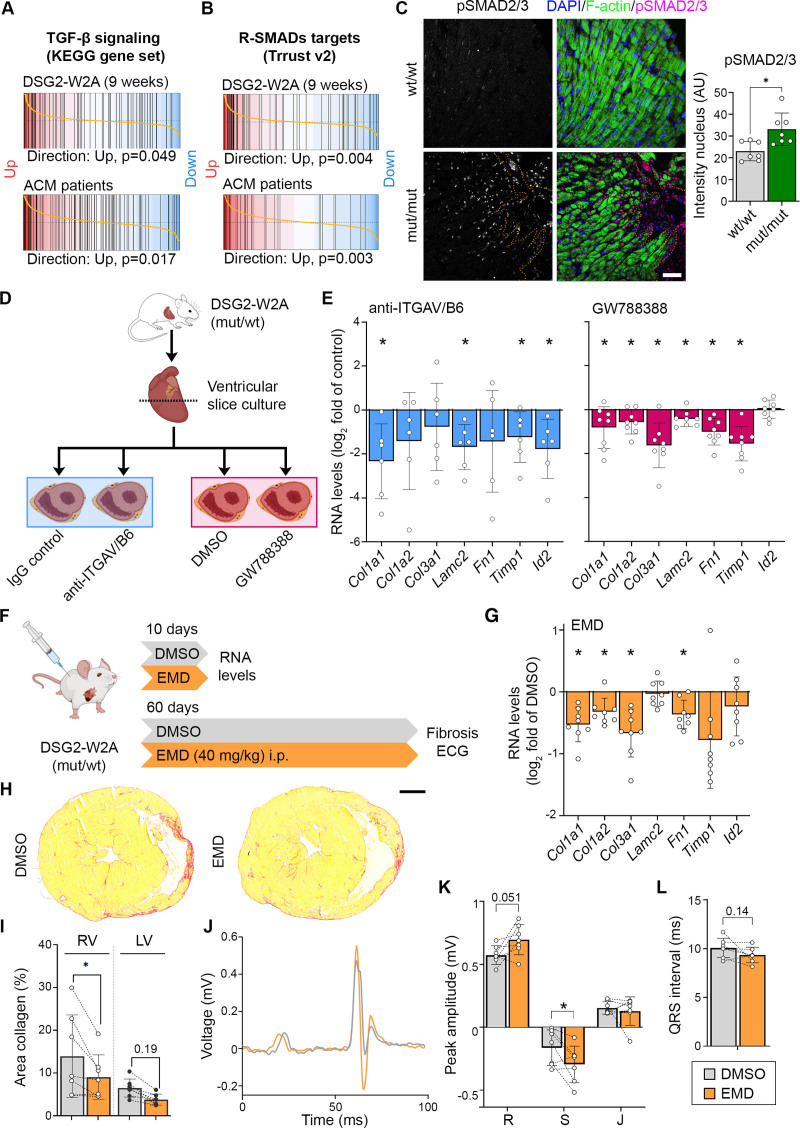
Figure 6.
Elevated transforming growth factor–β signaling in DSG2 (desmoglein-2)–W2A hearts as result of ITGAV/B6 activity. Barcode plots of gene set enrichment analysis of (A) the KEGG_TGF_BETA_SIGNALING_PATHWAY data set (systematic name: M2642)41 and (B) genes directly regulated by receptor-regulated SMADs (R-SMADs; includes SMAD1/2/3/5/9) as published in the TRRUST database21 in 9-week-old DSG2-W2A (mut/mut vs wt/wt) or arrhythmogenic cardiomyopathy (ACM) patient data set 1 (ACM versus healthy control; GEO: GSE107157/GSE107480). Indicated P values are calculated by function cameraPR of the R package limma. C, Immunostaining of phosphorylated SMAD2/3 (magenta, S465/S467 or S423/S425, respectively) in sections of DSG2-W2A hearts and related analysis of nuclear staining intensity. Nuclei are stained with DAPI (blue), cardiomyocytes are marked with f-actin (green). The dotted orange line marks the edge of the fibrotic area. Scale bar, 50 µm. *P<0.05, unpaired Student t test. D, Schematic of experimental setup for integrin-αVβ6 (ITGAV/B6) blocking experiments in cardiac slice culture with related results in E. Icons are derived from BioRender. E, Reverse transcription quantitative polymerase chain reaction analysis of expression of genes downstream of transforming growth factor–β (TGF-β) signaling in cardiac slice cultures treated with inhibiting anti-ITGAV/B6 (1:15) or 10 µmol/L GW788388, an inhibitor of TGF-β receptor I, for 24 hours. *P< 0.05, paired Student t test versus indicated control condition. F, Schematic of experimental setup for in vivo ITGAV/B6 blocking experiments by injection of 40 mg/kg EMD527040 (EMD) intraperitoneally daily. DMSO was applied as vehicle control. Icons are derived from BioRender. G, Reverse transcription quantitative polymerase chain reaction expression analysis of genes downstream of TGF-β signaling in hearts of mice treated with EMD or respective amount of DMSO for 10 days. *P<0.05, unpaired Student t test versus DMSO. H and I, Cardiac fibrosis detected by picrosirius red collagen staining with representative images and corresponding analysis of the area of collagen in the right ventricle (RV) and left ventricle (LV). Lines indicate littermates. Each dot represents 1 animal. *P<0.05 or as indicated, grouped 2-way repeated-measures analysis of variance with LV/RV and experimental pairs matched, Sidak post hoc test. Scale bar, 1 mm. J through L, ECG recoded in lead II with representative curves shown in J. Corresponding analysis of R, S, and J peak amplitude and QRS interval, *P<0.05 or as indicated. Paired Student t test. Lines indicate littermates. Each dot represents 1 animal.