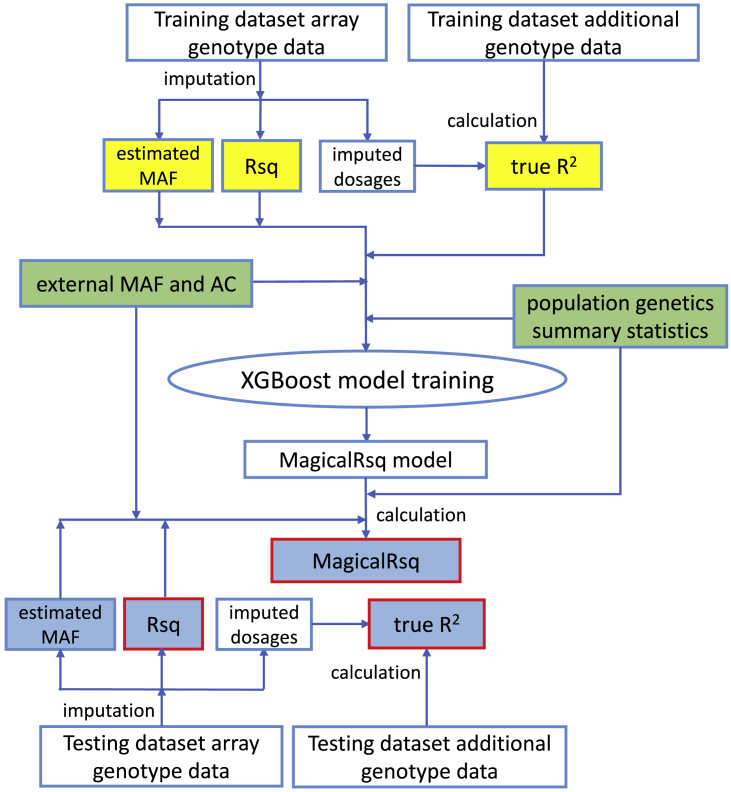
Figure 1.
MagicalRsq workflow
MagicalRsq starts from “training dataset array data” (which are data used for imputation among training individuals) and performs imputation using these data, which gives us standard Rsq and estimated MAF for each marker, in the training dataset. Then we calculate the true R2 by comparing imputed dosages with truth genotypes (established by additional genotype data in the training set). Combining external MAF and alternative allele count (AC), as well as population genetics summary statistics, with the above three metrics (i.e., standard Rsq, estimated MAF, and true R2), we train MagicalRsq models using the XGBoost method where we build supervised models to predict true R2 from all the other features. We then proceed to the testing dataset where we follow the same imputation workflow starting again from array genotype data and obtaining estimated MAF and standard Rsq after imputation. We then calculate MagicalRsq in the testing dataset by plugging in the predictor features into the MagicalRsq models built from the training dataset. Finally, we evaluate the performance of MagicalRsq (and Rsq) by comparing with true R2 in the testing dataset. Yellow highlights represent all the instruments specific for the training dataset, light blue highlights represent the instruments specific for the testing dataset, green highlights represent external information used in both training and testing, and red rectangles represent statistics used during final evaluation and comparison of MagicalRsq and standard Rsq, using true R2 as the gold standard.