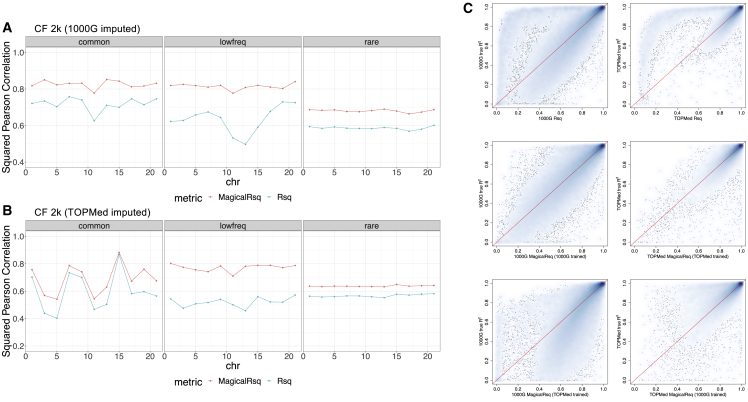
Figure 2.
Scenario 1, experiments 1–4: Training using even-numbered chromosomes and testing on odd-numbered chromosomes for CF 2k samples
(A and B) Performance comparison between Rsq and MagicalRsq in terms of squared Pearson correlation with true R2 for (A) 1000G-based imputation; (B) TOPMed-based imputation.
(C) Smooth scatterplot showing Rsq or MagicalRsq (x axis) calculated from both matched- (second row) and mis-matched- (third row) models against true R2 (y axis) for both 1000G-based (left) and TOPMed-based (right) imputation, for low-frequency variants on chromosome 13.