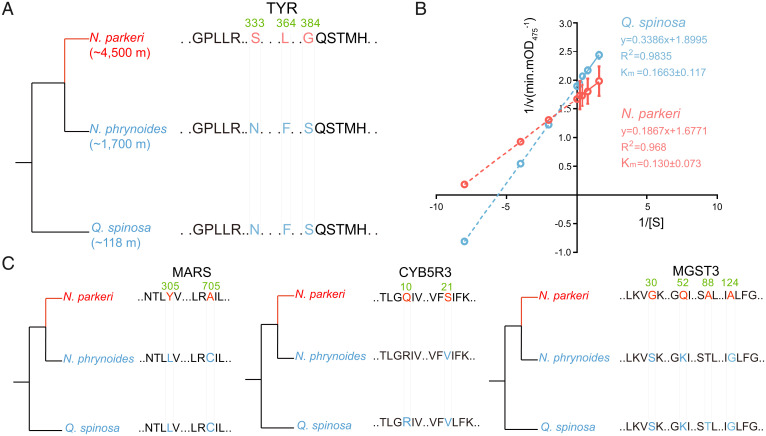
Fig. 4.
Amino acid replacements of PSGs and enzymatic activity identification of tyrosinase (TYR) across N. parkeri and Q. spinosa. (A) Phylogenetic relationships and schematic alignment of TYR amino acid sequences in N. parkeri (red) and its lower-elevation relatives (N. phrynoides and Q. spinosa; blue). Amino acid replacements unique to N. parkeri shown in red were found to be under positive selection. The reference alignment numbers refer to the amino acid sequence of N. parkeri. (B) In vitro enzymological kinetic analyses of TYR in N. parkeri and Q. spinosa. Km (Michaelis constant) is used to describe the rate of enzymatic reactions. It is numerically equal to the substrate concentration [S] at which the reaction rate is half of the maximum rate [v] achieved by enzyme. The smaller Km represents the stronger affinity the enzyme has for the substrate. Each Km value is derived from eight independent experiments, which are indicated with circles. R2 represents the correlation coefficient of the kinetic equation that fits the reaction. (C) Schematic amino acid alignments of MARS, CYB5R3, and MGST3 in N. parkeri (red) and its lower-elevation relatives (blue). Single-letter abbreviations for the amino acid residues are as follows: F, Phe; G, Gly; H, His; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; S, Ser; T, Thr; V, Val.