Figure 6.
Overview of the in vitro mutant versus wild-type luciferase assays in ARPE-19 cells
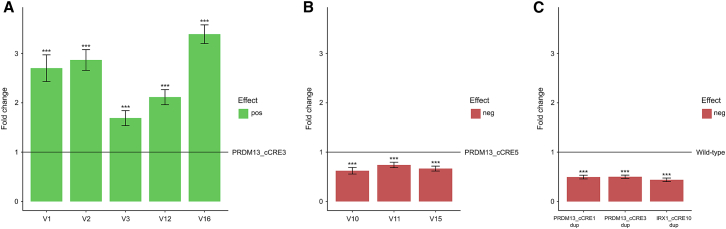
(A) In mutational hotspot-1 (PRDM13_cCRE3) upstream of PRDM13, the four known (V1–V3, V12) variants and the V16 variant demonstrate a significant increase of luciferase reporter activity (p < 0.001), relative to the wild-type vector.
(B) In contrast, the two known (V10, V11) variants and the V15 variant located in mutational hotspot-2 (PRDM13_cCRE5) upstream of PRDM13 cause a significant decrease of luciferase reporter activity (p < 0.001), relative to the wild-type vector.
(C) For three non-coding regions of interest, located in the shared duplicated region of either the PRDM13 or IRX1 locus, the level of luciferase reporter was reduced by half (p < 0.001) when the region was present as tandem duplication, relative to their respective wild-type counterpart, containing the same region of interest as a single insert.
neg, negative; pos, positive; ∗∗∗p < 0.001.