Abstract
Phospholipid bilayers are liquid-crystalline materials whose intermolecular interactions at mesoscopic length scales have key roles in the emergence of membrane physical properties. Here we investigated the combined effects of phospholipid polar headgroups and acyl chains on biophysical functions of membranes with solid-state 2H NMR spectroscopy. We compared the structural and dynamic properties of phosphatidylethanolamine and phosphatidylcholine with perdeuterated acyl chains in the solid-ordered (so) and liquid-disordered (ld) phases. Our analysis of spectral lineshapes of 1,2-diperdeuteriopalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE-d62) and 1,2-diperdeuteriopalmitoyl-sn-glycero-3-phosphocholine (DPPC-d62) in the so (gel) phase indicated an all-trans rotating chain structure for both lipids. Greater segmental order parameters () were observed in the ld (liquid-crystalline) phase for DPPE-d62 than for DPPC-d62 membranes, while their mixtures had intermediate values irrespective of the deuterated lipid type. Our results suggest the profiles of the acyl chains are governed by methylation of the headgroups and are averaged over the entire system. Variations in the acyl chain molecular dynamics were further investigated by spin-lattice () and quadrupolar-order relaxation () measurements. The two acyl-perdeuterated lipids showed distinct differences in relaxation behavior as a function of the order parameter. The R1Z rates had a square-law dependence on , implying collective mesoscopic dynamics, with a higher bending rigidity for DPPE-d62 than for DPPC-d62 lipids. Remodeling of lipid average and dynamic properties by methylation of the headgroups thus provides a mechanism to control the actions of peptides and proteins in biomembranes.
Significance
Lipid composition is critically involved in regulating membrane mechanical properties and cellular functioning. Different lipid components affect the properties of membranes, which may explain their biological diversity. Solid-state NMR spectroscopy shows that, in the liquid-disordered (liquid-crystalline) phase, the smaller headgroup size of phosphatidylethanolamines has striking effects on acyl chain packing and dynamics compared with phosphatidylcholines. Close packing of the phosphoethanolamine headgroups restricts the cross-sectional area of the acyl chains, yielding higher segmental order parameters and greater bilayer thickness. Solid-state NMR relaxation further informs dynamic lipid properties and indicates an increased bending rigidity of phosphatidylethanolamine bilayers versus phosphatidylcholines.
Introduction
Biological membranes are characterized by a strikingly large assortment of different lipid species (1, 2, 3) that may be related to the functions of their protein constituents. Variation of the lipids can have wide-reaching effects over the entire membrane structure on account of the highly collective intermolecular interactions. It has become well established that the lipid environment can substantially alter the functions and energetics of membrane proteins (4, 5, 6, 7, 8, 9) through the involvement of both the lipid polar headgroups and acyl chain substituents (3,10, 11, 12, 13). One hypothesis is that lipids control membrane protein activity by allosteric modulation due to binding to specific recognition sites, as for diacylglycerol and protein kinase C (14,15), viral membrane proteins (16), and G-protein-coupled receptors (GPCRs) (17,18). Alternatively, the interactions may be nonspecific and entail modification of physical properties of the entire bilayer, including phase behavior, membrane thickness, molecular packing, surface charge density, lipid shape (19,20), and curvature stress (7). Recent literature has confirmed the asymmetric segregation of membrane lipids such as phosphatidylethanolamine (PE) and phosphatidylcholine (PC) in cellular plasma membranes, with PE being much more prevalent in the inner leaflet while PC is localized to the outer leaflet (3), which may affect properties such as the bending rigidity (21). A further important aspect is whether natural selection of membrane properties has occurred for lipids that may act as regulators of membrane protein structure and activity. Investigations of the chemical and physical properties of the major cellular lipid constituents such as PE and PC are thus highly significant to understanding how protein functions are modulated by the lipid diversity presented in biomembranes (3,22).
To explore these questions, solid-state deuterium (2H) nuclear magnetic resonance (NMR) spectroscopy has been applied to biomembranes (23, 24, 25, 26, 27) and used to study the configurational properties and molecular dynamics (21,28, 29, 30) of phospholipid bilayers at the atomistic level. Investigations of the solid-ordered (so) and liquid-disordered (ld) phases of membrane lipids have been carried out (31, 32, 33, 34) together with studies of liquid-ordered (lo) raftlike lipid mixtures (21,35,36). In these cases, the physical properties include effective acyl chain lengths and average chain cross-sectional areas, as well as moduli for elastic distortions of phospholipids, which can be interpreted by simple statistical models (37). As an example, the mean area per lipid molecule at the membrane-water interface in the ld state (33,37) is essential to validating molecular dynamics (MD) simulations (38, 39, 40). Likewise, the bending rigidity and spontaneous curvature have crucial roles for relating the curvature free energy of lipid membranes to their functional mechanisms (41). Such material parameters describe the properties of biological membranes that can modulate the structure and activity of peptides and proteins or the lipid bilayers themselves.
Here we address the hypothesis that combination of lipid polar headgroups and nonpolar acyl chains controls cellular function through modulation of bilayer properties in the liquid-crystalline (Lα) state (Fig. 1 A and B). We characterized influences of methylation of the polar headgroups in remodeling the hydrocarbon core of lipid bilayers, with emphasis on the structure and dynamics of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) model bilayers. Because the acyl chains are identical, any differences are clearly due to the polar headgroups. Solid-state 2H NMR spectra were acquired of binary mixtures of DPPE and DPPC in the so phase and ld phase with the acyl chains of one or the other component perdeuterated, and analyzed by moments of the spectral lineshapes and order parameters. We discovered that methylation of phosphoethanolamine headgroups to yield phosphatidylcholine has striking effects on material properties of the bilayer hydrocarbon core. In the ld phase, mean-torque analysis of the order profiles further showed that membranes with phosphoethanolamine headgroups are thicker, with a smaller area per lipid versus methylated phosphocholine substituents. Model-free analysis of nuclear spin relaxation rates revealed how the phospholipid headgroups control membrane deformation. Bringing together the solid-state 2H NMR order parameters and relaxation times indicates PE membranes have higher bending rigidities than PC bilayers over mesoscopic length scales, while mixtures with different lipid headgroups show intermediate structural and dynamic properties. Our results inform how cellular membranes can regulate protein structure and activity through the chemically nonspecific balance of acyl chains and polar headgroups (7). The methods highlighted here are furthermore significant to validating MD simulations of lipid membranes (39,42,43), sterols (21), and proteins (44,45) in biomembranes.
Figure 1.
Hierarchical dynamics of phospholipid bilayers influence material properties, including the interfacial area per molecule, bilayer thickness, and monolayer bending rigidity. (A) Transformations and dynamic time scales involved in solid-state 2H NMR spectroscopy showing local and collective fluctuations of lipid membranes. Euler angles (Ω) are illustrated with subscripts denoting the various rotational transformations. Coordinate systems are defined as follows: I, internal or intermediate segmental frame; M, molecular interaction frame; D, bilayer director frame; and L, laboratory frame. The z-axes of the coordinate frames are indicated. (B) Illustration of how volumetric data, hydrocarbon thickness DC, and mean cross-sectional chain area ⟨AC⟩ correspond to the average area ⟨A⟩ per lipid molecule in the bilayer. For a polymethylene chain, a three-carbon segment (virtual bond) is defined from carbon Ci−1 to Ci+1 with length DM = 2.54 Å and projection ⟨Di⟩ onto the molecular (M) axis. The orientation of the z-axis of the intermediate (I) frame to the M frame is shown. Structural parameters for Lα membranes are derived from solid-state 2H NMR spectral data by applying a mean-torque model (37). Figure adapted from Refs. (25,26). To see this figure in color, go online.
Materials and methods
Fully protiated DPPE and DPPC were obtained from Avanti Polar Lipids (Alabaster, AL), and were used without further purification. Organic synthesis of 1,2-diperdeuteriopalmitoyl-sn-glycero-3-phosphocholine (DPPC-d62) was carried out by acylating the cadmium chloride adduct of sn-glycero-3-phosphocholine (prepared from locally obtained hen egg yolks) with the anhydride of palmitic acid-d31 (46). The 1,2-diperdeuteriopalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE-d62) was synthesized by transphosphatidylation of the DPPC-d62 using phospholipase D (from locally obtained Savoy cabbage) in the presence of ethanolamine. All lipids were purified using silica gel column chromatography, and yielded a single spot with thin-layer chromatography upon elution with CHCl3:MeOH:H2O (6:4:1) and visualization by exposure to I2 vapor and/or by spraying with 40% H2SO4 in ethanol and charring at 270°C. The DPPE-d62 and DPPC-d62 samples were dried under high vacuum, placed in 8-mm diameter test tubes (Schott Glass, Jena, Germany), and mixed with 50 wt % buffer containing 20 mM MOPS and 1 mM EDTA in 2H-depleted water at pH 7.1. All lipid mixtures were prepared by combining appropriate amounts in chloroform:methanol (2:1), evaporating the solvent in an 8-mm test tube, and mixing with 50 wt % buffer. Samples included at least 10 mg of acyl-chain-perdeuterated lipids to obtain an acceptable signal-to-noise ratio. The resulting multilamellar dispersions were then annealed by multiple freeze-thaw cycles.
Solid-state NMR spectroscopy of multilamellar lipid dispersions
The 2H NMR experiments were conducted at a magnetic field strength of 7.06 Tesla (2H frequency of 46.1 MHz) utilizing a home-built, high-power horizontal solenoid probe with an 8-mm diameter radiofrequency coil, an external digitizer, and a tuned high-power radiofrequency amplifier (Henry Radio Tempo 2006-A). Hydrated lipid samples were contained in sealed, cutoff 8-mm test tubes placed within the radiofrequency coil. An eight-step, phase-cycled, quadrupolar-echo pulse sequence, , was used for data acquisition. For the lineshape experiments, a relatively short 2-μs pulse, with a pulse spacing of 40 μs, a 2-μs dwell time, and a recycle time of 500 ms were used. Free-induction decays were recorded with both increasing and decreasing temperatures, and samples were equilibrated at each temperature for approximately 30 min before data acquisition. A typical quadrupolar-echo spectrum took approximately 1 h to acquire, and each relaxation experiment involved about 12 h of signal acquisition. The spectra were obtained by Fourier transformation using both quadrature channels beginning precisely at the top of the quadrupolar echo. Moment analyses and de-Pakeing as introduced by Bloom et al. (47) were carried out with modified versions of the programs of Refs. (47) and (48). The solid-state 2H NMR lineshape simulations used the program MXQET (49), which allows for multisite, multiaxis motional models. The simulations (six-site jump) corresponded to diffusion about the long axis of the molecule. The 2H NMR spectra of the methylene segments and the methyl groups were calculated separately, scaled by the number of deuterons, and added to yield the simulated powder patterns. The experimental 2H NMR spectra were fitted by adjusting the jump rate between the sites, the effective quadrupolar coupling constant, and the degree of Lorentzian or Gaussian line broadening. The jump rate (kjump) corresponds to the axial diffusion constant (DR) by , where is the angle between the sites, which is 60° for the six-site jump motion.
Spin-lattice relaxation times (T1Z) were acquired using a 32-pulse, phase-cycled inversion-recovery sequence (50), and quadrupolar-order relaxation time (T1Q) results were obtained with a composite broadband pulse sequence (51). Relaxation experiments were obtained with a quadrupolar-echo pulse spacing of 30 μs, a dwell time of 7 μs, and a recycle delay of 800 ms. The relaxation rates were obtained for each of the resolved peaks in the de-Paked 2H NMR spectra. Nonlinear regression fits of the recovery curves to the functions or were used to obtain the spin-lattice relaxation (R1Z) rates or quadrupolar-order relaxation (R1Q) rates, respectively, where M0 is the intensity at equilibrium, and incompleteness of the magnetic inversion is accounted for by the W parameter. The de-Pakeing algorithm assumes the 2H NMR powder-type spectra are axially symmetric and scale as , where P2 is the second-order Legendre polynomial and θ is the angle of the bilayer normal to the static magnetic field (48). Orientational anisotropy of the relaxation rates (52) in principle leads to changes in the lineshape as a function of the variable time delay in both the spin-lattice and quadrupolar-order relaxation experiments. However, rapid orientational averaging occurs on the time scale of relaxation experiments in phospholipids such as DPPC, as first shown by Brown and Davis (53). Motional averaging yields axially symmetric powder-type spectra whose shape is unaffected by the variable delay time. We assumed that similar behavior exists for the DPPE-d62 multilamellar dispersions. Hence, although the de-Paked subspectra correspond to the θ = 0° bilayer normal direction, the relaxation rates are an average over all orientations.
Segmental order parameters and moments of spectral lineshapes
The solid-state 2H NMR spectra reveal a distribution of quadrupolar splittings that manifest the equilibrium or average properties of the membrane lipids. For lipids in a bilayer arrangement, the motional averaging axis is the membrane normal, and the shape of the 2H NMR spectra corresponds to axially symmetric motion. For such a case, the residual quadrupolar splitting between the two spectral transitions for a given labeled segment (i) is related to the C−2H bond segmental order parameter () by (27).
| (1) |
Here is the static quadrupolar coupling constant and is approximately 170 kHz for a methylene C−2H bond (54), P2 is the second-order Legendre polynomial, and θ is the angle between the bilayer normal (director axis) and the static magnetic field. The orientational order parameter for the ith segment is defined as
| (2) |
where is the angle of the C−2H bond of the ith segment to the bilayer normal and the angular brackets indicates a time or ensemble average (27). The observed order parameters were derived using Eq. 1 from the de-Paked solid-state 2H NMR spectra.
Analysis by spectral moments is extremely useful, as introduced by Davis (55), because it yields information on the overall lineshape even when individual splittings are not resolved. The first and second half-moments of the solid-state 2H NMR spectra were calculated from the experimental lineshape distribution function f(ω) (56) by
| (3) |
In the above expression, k = 1 or 2, and f(ω) is the intensity of the spectral lineshape at a frequency ω away from the center frequency (ω = 0). In terms of the order parameters, the expressions for the first and second moments read:
| (4a) |
| (4b) |
Additionally, the distribution of order parameters about the mean is characterized by the parameter Δ2, defined as (55).
| (5a) |
| (5b) |
which corresponds to the fractional mean-squared width or variance. The above expressions characterize how the 2H NMR lineshapes are related to the distributions of residual quadrupolar splittings (RQCs); i.e., order parameters averaged over the lipids as indicated by the angular brackets.
Calculation of average membrane properties using mean-torque model
Further interpreting the |SCD| order parameters in terms of structural quantities entails introduction of various motional models (37,57). In previous work, a simple diamond-lattice model was used for the configurational statistics of the polymethylene acyl chains of lipid bilayers in the ld phase (56). However, such an approach may unduly restrict the segmental distribution and overestimate the area per lipid at the aqueous interface (58). An alternative is a mean-torque potential model (37) to connect the order parameters with the lipid nanostructure. As introduced by Petrache et al. (37), this model is impactful for validating MD simulations of lipid bilayers (38,59). For single-component bilayers, or when the membrane composition is mixed, the interactions between neighboring molecules can change the average cross-sectional area per lipid. At the molecular scale, on average each lipid in the membrane occupies a space related to the volume and length of the hydrocarbon chains by
| (6) |
where DC is the volumetric thickness of an individual monolayer, and VC is the total volume of an individual acyl chain. In Eq. 6, the chain volume VC is given by the densitometry measurements of Nagle et al. (60), and is conserved (i.e., constant). For diacyl lipids, the interfacial area per molecule is given by ⟨A⟩ = 2⟨AC⟩ in terms of the area of an individual chain (37,61). Notably, the volumetric thickness DC and the mean area per lipid 〈A〉 are inversely related, meaning the bilayer core has nearly the density of liquid hydrocarbon (62). Hence, the volumetric thickness DC is not the same as the mean projected acyl length (56). Because of end effects of the acyl chains, the mean travel away from the aqueous interface is less than the distance to the bilayer midplane (37).
To avoid complications from chain upturns (37), the relatively ordered acyl segments near the headgroup can be used instead of the full hydrocarbon chain. At a certain bilayer depth, the influence of chain terminations becomes significant (63). Acyl chains on adjacent molecules are more disordered beyond this point to maintain the packing at hydrocarbon density (62). The largest order parameter corresponds to the so-called plateau region of the 2H NMR order profiles, where the segmental cross-sectional area and projected length are inversely correlated (56). As first pointed out by Jansson et al. (56), the average cross-sectional area per lipid is given as (37)
| (7) |
The reader should note that because of the distribution of segment orientations. Additionally, is the methylene volume (60), and D the instantaneous travel of an individual segment along the bilayer normal. Following Petrache et al. (37), the above expression can be recast as
| (8) |
in which DM = 2.54 Å is the maximum projection onto the bilayer normal of the virtual bonds connecting every second polymethylene chain carbon, and is the lipid cross-sectional area in the all-trans conformation (37). The area factor q is defined as where βi is the angle of the virtual bond between the two Ci–1 and Ci+1 carbon atoms and the normal to the lipid bilayer surface (64).
In Eq. 8, the shape of a statistical segment is approximated by a geometrical prism with a constant hydrocarbon volume. The effective acyl segment length is averaged over the motions, while the segmental volume is conserved. For a given acyl segment (index i) a Taylor series expansion about the all-trans value allows the area factor q in Eq. 8 to be approximated by up to the second order (37). Using solid-state 2H NMR spectroscopy, the second moment is then obtained directly from the segmental order parameters by
| (9) |
Calculation of the first moment for a given value of requires introduction of the potential of mean force, as described below.
The mean-torque model assumes the orientational order parameter for each chain segment relative to the local director is described by an orientational potential U(β) (potential of mean torque). The probability of finding a statistical segment with a virtual bond orientation β at any given instant is given by the Boltzmann distribution , where the chain index (i) is omitted. Here the partition function reads . Assuming a first-order mean-torque model (37), we find that U(β) = U1(cos β), where U1 is the first moment of the function U(β) in terms of Legendre polynomials. The angular-dependent quantities are integrated with the distribution function to give the coupled equations and . An analytical solution for is then found by using , which, for an individual segment (index i), yields
| (10) |
Take note that, for an all-trans conformation of the lipids, and hence q = 1. The volumetric (Luzzati) bilayer thickness is thus , where is the lipid volume (65,66). Eqs. 7 and 8 for an all-trans chain give a limiting area of and limiting monolayer thickness of , where is the number of acyl chain carbons. The effective membrane thickness is likewise calculated using , where is the volumetric hydrocarbon thickness of the acyl chains of the lipids and is the headgroup plus the backbone distance. In the case of phospholipids, is 9 Å (67,68). The values of DH plus DC yield the steric bilayer thickness as defined by Nagle et al. (65). Following Petrache et al. (37), the chain volume at temperature T is found from the methylene volume using , where is the thermal expansion coefficient for methylene groups (20,60,69).
Analysis of nuclear spin relaxation rates in terms of irreducible spectral densities of motion
In lipid bilayers, the molecular motions cause time-dependent fluctuations in the quadrupolar Hamiltonian, giving the relaxation observed in solid-state 2H NMR spectroscopy (25,26). For a stationary Markoff process, the irreducible correlation functions Gm(τ) describe the fluctuations of the C−2H bond orientation (i.e., electrostatic field gradient tensor (EFG)) at a time t relative to its value at a time t + τ later. The indices m = 0, ±1, and ±2 are for projection of the nuclear spin angular momentum onto the axis of quantization (magnetic field). Coupling of the 2H nuclear quadrupole moment with the EFG of the C−2H bond gives a time-dependent perturbation within the interaction picture. If we know the motion, the correlation function can be deduced and vice versa. Fourier transformation of Gm(τ) then yields the spectral densities of motion, denoted by (70), which read as follows:
| (11) |
where ω0 is the Larmor frequency. The spectral densities (power spectra) are directly related to the solid-state 2H NMR relaxation rates (71), as given by
| (12a) |
| (12b) |
Through measuring both the R1Z and R1Q rates, the irreducible spectral densities of motion and can be experimentally determined (e.g., as input for all-atom MD simulations). Notably, translational diffusion of the lipids in multilamellar dispersions leads to orientational averaging of the relaxation rates, where the dependence on the projection index (m) vanishes due to the spherical symmetry, giving for the orientationally averaged spectral densities. For additional explanation see Refs. (25,26).
Results
Phospholipid headgroups govern bilayer structure in the solid-ordered phase
For multilamellar dispersions of DPPE-d62 in the so phase (or gel, Lβ phase), the solid-state 2H NMR spectra comprise broad powder patterns. Two distinct splittings are evident at = ±4.5 kHz and ±23 kHz at 42°C (Fig. 2 A). However, these splittings are less than expected for the methyl and methylene groups of an all-trans chain undergoing axial rotation ( = ∓10.6 and ±32 kHz, respectively). With temperature reduction, the experimental spectra change further (Fig. 2 B), obscuring the splittings at ±23 kHz. Particularly the solid-state 2H NMR spectra of DPPE-d62 bilayers acquired at 29°C resemble those for DPPC-d62 at nearly the same temperature (Fig. 2 C). However, the DPPC-d62 bilayer yields this type of spectra at all temperatures below the main order-disorder phase transition (55,72). Although the methylene segments do not appear completely all-trans, they are roughly equivalent. To further interpret the data, spectral lineshape simulations were carried out (Fig. 2 A−C). For all three cases, an effective coupling constant of ≡ 150 kHz was used to fit the spectral data. The simulations indicated a nearly all-trans configuration for the acyl chains, with a whole-molecule wobbling motion (order parameter S ≈ 0.9) that further averages the EFG tensors (Fig. 2 D) to give the experimental quadrupolar splittings. The differences for DPPE-d62 at 42°C (Fig. 2 A) and 29°C (Fig. 2 B) come mainly from the rate of axial diffusion, which is 6.58 × 106 rad s−1 for the 42°C and 2.85 × 106 rad s−1 for the 29°C simulations. The simulated spectra for DPPE-d62 at 29°C and for DPPC-d62 at 31°C are nearly identical, except for slight changes in the amount of line broadening. In both cases, the so phase shows significant mobility and disorder of the lipids at the molecular level. For DPPE bilayers the molecular packing is tighter and molecular order is higher, whereas, in DPPC, the greater motional averaging yields smoother shoulders requiring additional line broadening to simulate the lineshapes.
Figure 2.
Solid-state 2H NMR spectra for DPPE-d62 and DPPC-d62 multilamellar dispersions in the low-temperature (solid-ordered (so)) state reveal internal chain mobility. (A) The solid-state 2H NMR spectrum of DPPE-d62 immediately below the order-disorder phase transition indicates the methylene groups of the acyl chains are nearly equivalent and experience rapid axial rotation yet are not in the all-trans state. (B) When temperature is lowered, the 2H NMR spectra lose the distinct splittings at ±23 kHz. (C) The low-temperature spectrum for DPPC-d62 below the order-disorder phase transition likewise shows the chains are not completely all-trans and rotate axially. Dotted lines over the spectra show the simulated solid-state 2H NMR spectra. Simulations in (A) and (B) differ only by the rate of axial rotation, while (B) and (C) differ only in the line broadening. (D) Simulated 2H NMR lineshape after introducing lipid rotation about the director axis. To see this figure in color, go online.
Changes in the first and second moments of the solid-state 2H NMR lineshapes with temperature (Fig. 3 A and B) strikingly reveal the order-disorder phase transitions of the two lipids, with very little hysteresis. The main phase transition temperature, Tm, for DPPE-d62 is approximately 20°C higher than DPPC-d62 as shown by the mean-square deviation of the order parameters (Fig. 3 C) (Tm = 56°C versus 38°C; note the 2H isotope effect). Under these conditions, both phospholipids are in the lamellar phase, because the transition to the nonlamellar (HII) phase occurs at appreciably higher temperatures (118°C for DPPE-d62) (73). The moments for the phosphatidylcholine bilayer are less than the phosphatidylethanolamine bilayer between 30°C and 37°C, which may indicate another phase, such as the Pβ′ ripple phase with tilted chains (74,75). This pretransition has been observed by differential scanning calorimetry, and occurs from the Lβ′ phase, in which the acyl chains are all-trans and tilted with respect to the bilayer normal (76), to the Pβ′ ripple phase (73). Below the pretransition, for DPPC-d62 the moments continue to increase with decreasing temperature (Fig. 3 A and B). The pretransition is not seen for DPPE-d62 using solid-state 2H NMR, nor is it observed by differential scanning calorimetry (77). We thus conclude that, in the so (gel) phase, both DPPE and DPPC lipids are nearly all-trans, although rotating about their long axes gives additional disorder.
Figure 3.
Moment analysis of solid-state 2H NMR spectra of multilamellar lipids reveals structural information for solid-ordered (so) and liquid-disordered (ld) states. (A) First moment M1, (B) second moment M2, and (C) mean-squared deviation Δ2 are shown as a function of increasing (filled symbols) and decreasing (open symbols) temperature for DPPE-d62 and DPPC-d62 multilamellar dispersions. The large change in moments is due to the main chain melting transition, which occurs at 56°C for DPPE-d62 and 38°C for DPPC-d62 bilayers. Neither sample shows appreciable hysteresis in the phase transition region. To see this figure in color, go online.
Structural properties of phospholipids are affected by polar headgroups in the liquid-disordered phase
The spectral moment analysis establishes how the multilamellar DPPE-d62 and DPPC-d62 lipids undergo the phase transition to the ld phase (or Lα phase) as the temperature is increased. We now consider the biologically relevant liquid-crystalline state in greater detail, as investigated by solid-state 2H NMR spectra of DPPE-d62 and DPPC-d62 in the ld phase (Lα phase) (Fig. 4). Representative powder-type 2H NMR spectra of random multilamellar dispersions are shown at the top (Fig. 4 A and B), and oriented subspectra (θ = 0°) obtained by numerical deconvolution (de-Pakeing) (47) are given at the bottom (Fig. 4 C and D). From the de-Paked subspectra, the distribution of RQCs is clearly evident. The largest splitting corresponds to the acyl segments near the polar headgroups, with a progressive decrease until the terminal methyl groups with the smallest splittings are reached (27). At nearly the same absolute temperature, the residual quadrupolar splittings are clearly greater for DPPE-d62 than for the DPPC-d62 bilayer (Tables S1 and S2), and more methylene groups contribute to the maximum splitting giving greater intensity. This behavior is also reflected in the spectral moments, which are higher for DPPE-d62 compared with DPPC-d62 at the same temperature (Fig. 3). Hence, in the ld phase, the ordering of the acyl chains is greater for DPPE-d62 than for the DPPC-d62 bilayers. It immediately follows that, in the ld phase, the DPPE bilayer has a smaller polar headgroup area at the aqueous interface and a correspondingly greater bilayer thickness (37).
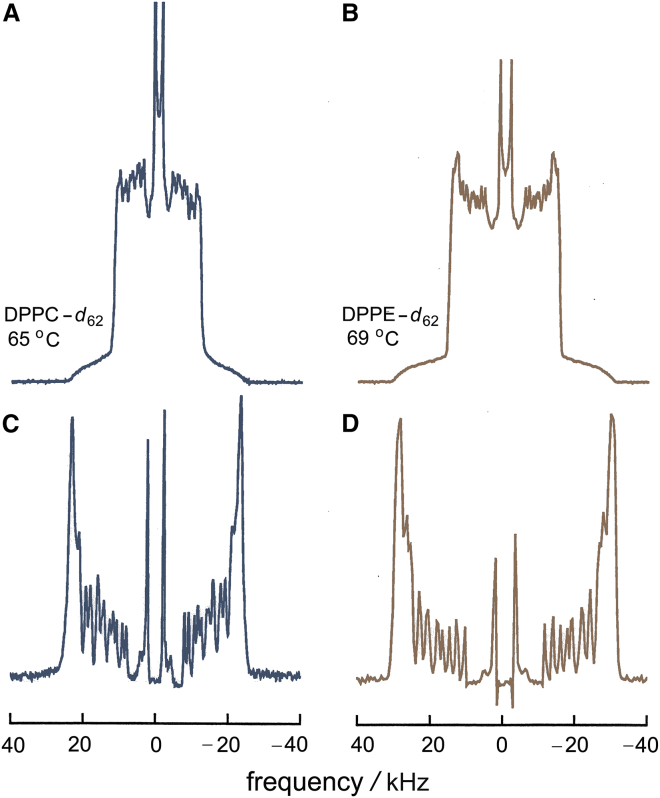
Figure 4.
Solid-state 2H NMR spectra of multilamellar lipid dispersions show distributions of residual quadrupolar splittings (RQCs) in the ld or Lα phase. Representative data are shown for (A) DPPC-d62 at 65°C and (B) DPPE-d62 at 69°C in the Lα state. Powder-type 2H NMR spectra due to a random distribution of lamellae (A and B) are shown with the corresponding de-Paked spectra (C and D) directly beneath. The different methyl intensities in (C) and (D) arise from the different pulse sequence delays and are not significant. Both lipids give similar spectra, although the quadrupolar splittings for DPPE-d62 are larger compared with DPPC-d62 bilayers. To see this figure in color, go online.
Influences of PE headgroups on the acyl chains are quantitatively established from comparing plots of the segmental order parameters as a function of acyl carbon position (i) (Fig. 5 A and B) for multilamellar dispersions of DPPE-d62 and DPPC-d62 in the ld phase (Lα phase) at different temperatures (cf. Tables S1−S4). As evident from the moment analysis, the order parameters are higher for DPPE-d62 than DPPC-d62 at all temperatures, and the length of the plateau constant region is longer. This finding is not restricted to the palmitoyl acyl chains of DPPE-d62 and DPPC-d62. Similar behavior has also been observed when comparing bilayers of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine (DMPE) (78,79), as well as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), in the ld phase (Lα phase) (80). In these cases, the phosphatidylcholines showed smaller order parameters than the corresponding phosphatidylethanolamines at the same absolute temperature. Analogous results have been obtained by Fourier-transform infrared spectroscopy (FTIR) of DPPE-d4 specifically deuterated at the C4 acyl position, which shows a decrease in the fraction of gauche isomers compared with DPPC-d4 in the ld phase (Lα phase) (81). Higher values of the average bilayer thickness and a smaller average area per lipid are calculated for DPPE compared with DPPC using the mean-torque model (37,64), indicating the molecular packing is more condensed (Table S5). The observed molecular packing and bilayer thickness thus reflect the effect of methylation of the polar headgroups on the dynamic and material properties of the bilayers, as described below.
Figure 5.
Order parameter profiles of phospholipid acyl chains reveal differences due to phosphocholine and phosphoethanolamine headgroup substituents. (A) DPPE-d62 and (B) DPPC-d62 order profiles as a function of chain position (index i) are compared at various absolute and reduced temperatures in the ld or Lα phase. The smaller and larger values for the carbon segments arise from the sn-1 and sn-2 chains, respectively. Note the segmental ordering is greater for DPPE-d62 at all absolute and reduced temperatures compared with the DPPC-d62 bilayer. The estimated error in the order parameters is ±0.001. To see this figure in color, go online.
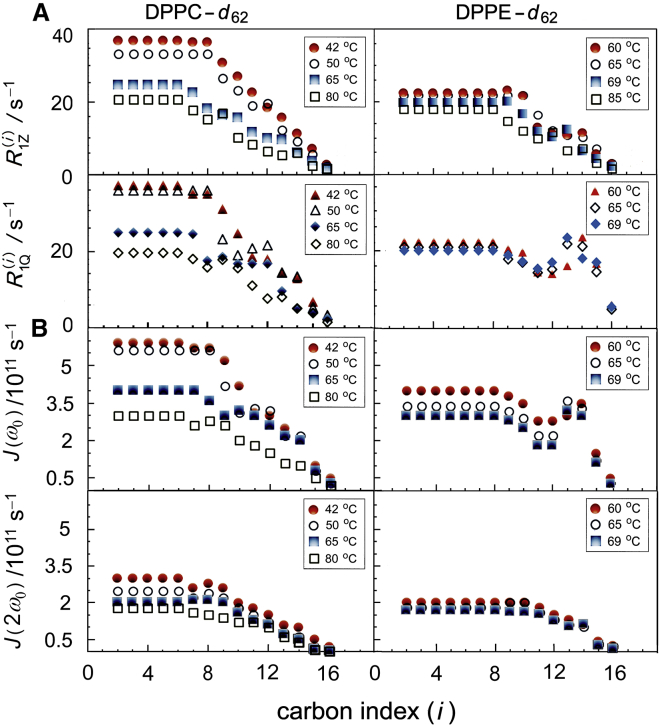
Further information on the phospholipid molecular dynamics can be experimentally obtained from solid-state 2H NMR relaxation measurements (25,26,82). A summary of our spin-lattice (R1Z) and quadrupolar-order (R1Q) relaxation rate studies for DPPE-d62 and DPPC-d62 multilamellar dispersions in the ld phase (Lα phase) is shown (Fig. 6 A) at different temperatures. The relaxation rates as a function of the carbon position (Tables S6 and S7) were measured from the de-Paked solid-state 2H NMR spectra. We assumed that averaging over all bilayer orientations occurs for DPPE-d62 as shown previously for DPPC-d62 bilayers (53), so that the de-Pakeing procedure is valid. Particularly the R1Z and R1Q relaxation rates for the DPPC-d62 multilamellar dispersions are significantly greater than for DPPE-d62 for a given |SCD| value at the same absolute temperature (Fig. 6 A). If comparison is made at the same reduced temperature Tred = (T − Tm)/Tm where Tm is the chain melting temperature (see above), the differences between DPPE-d62 and DPPC-d62 bilayers are even more pronounced. For both lipids, the relaxation rates diminish with greater temperature (Fig. 6 A), as seen for the homologous series of phosphatidylcholines (83). No maximum is found in either R1Z or R1Q (i.e., minimum in T1Z or T1Q). Hence, the motions causing the relaxation are fast with respect to the nuclear resonance (Larmor) frequency (>3 × 108 s−1). Additionally, for DPPC-d62, the relaxation rates decrease moving down the acyl chain (smaller |SCD| values), as observed for other phosphatidylcholines (83,84). This trend is also observed for the R1Z relaxation rates of DPPE-d62, although the R1Q rates decrease less as the order parameter is reduced. For DPPC-d62, the R1Z and R1Q relaxation rates are nearly equal for each observed |SCD| value and temperature investigated. However, the DPPE-d62 bilayers have a different behavior, as the R1Z and R1Q rates are equal only near the headgroup (high SCD; carbons ∼C2–C12). Clearly PE condenses the bilayer surface, although not as much as cholesterol, so that the smaller size and intermolecular hydrogen bonding of the polar headgroups yield greater bilayer thickness and reduced hydration versus PC bilayers. Variations also occur in the dynamic properties of the lipids due to their collective interactions within the membrane bilayers, which are discussed subsequently.
Figure 6.
Solid-state 2H NMR relaxation rate profiles of phospholipid acyl chains exhibit significant differences due to polar headgroups. (A) Profiles of relaxation rates and plotted against chain position (i) for multilamellar dispersions of DPPE-d62 and DPPC-d62 at various temperatures in the ld or Lα phase. Note the relaxation rates are smaller for DPPE-d62 than for the DPPC-d62 bilayer. (B) Corresponding profiles of model-free irreducible spectral densities of motion and calculated from experimental 2H NMR relaxation rates. The estimated error in the relaxation rates and spectral densities is ±5%. To see this figure in color, go online.
We next address the question of how we can disentangle the types, rates, and amplitudes of the lipid motions through combined measurements of the nuclear spin relaxation rates and the segmental order parameters (85). Here, the relaxation rate profiles are reminiscent of the order profiles, in that an approximate plateau is evident over the top part of the chain, decreasing with greater depth in the bilayer (Fig. 6 A). The results show an accumulation of the motional rates and/or amplitudes with greater depth within the hydrocarbon core. For all positions within the bilayer, the rates decrease with temperature, indicating the relevant motions have correlation times less than the inverse nuclear Larmor frequency (<2π/ω0 ≈ 3.4 ns). An additional important aspect is that, by measuring both the R1Z and R1Q rates, the irreducible spectral densities of motion J1(ω0) and J2(2ω0) are determined from Eqs. 12a and 12b as model-free experimental observables. Similar to the relaxation rates, the irreducible spectral densities decrease with greater temperature and smaller order parameters (Fig. 6 B). Notably, the NMR relaxation rates are averaged over all director orientations by translational diffusion of the lipids over the curved bilayer surfaces in multilamellar dispersions, as shown by the spectral hole-burning experiments of Brown and Davis (53). For the DPPE-d62 and DPPC-d62 bilayers, the orientationally averaged spectral densities are designated by ≡ , where, due to the spherical symmetry, the dependence on the projection indices m = 1, 2 with respect to the axis of quantization (magnetic field direction) vanishes. However, in this paper, we also retain the angular momentum projection indices (m = 1, 2) for generality.
Solid-state NMR spectroscopy characterizes structure and dynamics of lipid mixtures
Turning now to mixtures of phospholipids, their average and dynamic properties in the ld phase (Lα phase) can be similarly explored. The segmental order parameters for a 1:1 mixture of either DPPE-d62:DPPC or DPPC-d62:DPPE (Fig. 7 A) are intermediate to those of the pure DPPE-d62 or DPPC-d62 multilamellar dispersions (cf. Tables S3 and S4). We note that the order parameter profiles are essentially identical, regardless of which lipid species is deuterated. Such a situation has also been encountered with mixtures of DPPC and the corresponding lysolipid, 1-palmitoyl-sn-glycero-3-phosphocholine (PalPC) (56). These observations suggest that the order parameter is characteristic of the entire system and not the individual molecules in the (binary) mixture. The axial symmetry of the solid-state 2H NMR spectra about the bilayer normal (director) likewise implies that the order parameter is a collective property of the lipid nanostructure. Hence, one can investigate the average bilayer properties on the relevant time scale of solid-state 2H NMR spectroscopy (<10−5 s) (27) regardless of the probe lipid.
Figure 7.
Summary of results of solid-state 2H NMR spectroscopy for DPPE:DPPC (1:1) binary phospholipid mixtures in the ld state. (A) Segmental order parameters profiles and (B) relaxation rates and profiles at various temperatures. Note that the order parameters and relaxation rates are nearly independent of which component of the binary mixture is deuterated; i.e., whether it is DPPE-d62 or DPPC-d62 that is observed in the binary mixture. Hence the position of the deuterated acyl chains in the binary lipid mixtures has little effect on the observed order parameters and relaxation rates. (C) Square-law dependence of relaxation rates on segmental order parameters at 65°C in the ld phase (Lα phase). Data are for multilamellar lipid dispersions (50 wt % water). Note that the segmental order parameters, relaxation rates R1Z and R1Q, and slopes of the square-law plots are similar irrespective of the deuterated lipid molecules in the binary mixture. The estimated error in the order parameters is ±0.001 and the error in the relaxation rates is ±5%. To see this figure in color, go online.
As in the case of the pure compounds, for the binary mixtures of perdeuterated lipids, the relaxation rates R1Z and R1Q can be studied in similar fashion (Figs. S1−S5). Plots of the relaxation rates R1Z and R1Q as a function of carbon index (i) for 1:1 mixtures of DPPE:DPPC (Fig. 7 A and B) show the relaxation rates are nearly the same regardless of the headgroup of the labeled lipid (Tables S8 and S9). Because of the large two-phase region observed for DPPE:DPPC mixtures (86), it is difficult to define a reduced temperature, and so the relaxation rates are only compared at the same absolute temperature. For the DPPE:DPPC (1:1) mixtures, the R1Z relaxation rates in the so-called plateau region (highest |SCD| values) are greater than for either the single-component DPPE-d62 or the DPPC-d62 bilayers at the same absolute temperature (Fig. S1). For the lipid mixtures in the ld (or Lα) phase, both the equilibrium and dynamic bilayer properties are thus an average of those of the component lipids.
To recapitulate at this stage, our experimental findings show that methylation of phosphoethanolamine to give phosphatidylcholine yields a greater area per lipid and a decrease in the bilayer thickness, corresponding to a reduction of absolute spontaneous monolayer curvature (7,87). The segmental order parameters and relaxation rates are nearly independent of which component of the mixture is deuterated; i.e., whether it is DPPE-d62 or DPPC-d62 that is observed in the binary mixture. Accordingly, the area per lipid and dynamic properties are averaged over the entire bilayer on the 2H NMR time scale (<10−5 s) regardless of the probe lipid (21). Changes in both the order profiles and relaxation rate profiles further indicate an effect of headgroup methylation on collective lipid dynamics, thereby imparting diminished stiffness and bending rigidity of the bilayer for PC versus PE lipids.
Discussion
Solid-state NMR spectroscopy clearly indicates a role of the lipid polar headgroups in modulating collective dynamics of the acyl chains in biomembrane function. Experimentally, it is found that membrane proteins are sensitive to both the phospholipid headgroups and the acyl chains (7), implying that combined interactions are likely to be involved (7,88). Besides allosteric modulation of membrane protein activity, nonspecific lipid-protein interactions due to bulk bilayer properties can be implicated in membrane function. Accordingly, we studied two phospholipids with different polar headgroups, but identical acyl chains, namely 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC). In particular, DPPC has three methyl groups on the headgroup nitrogen, whereas DPPE lacks these quaternary groups and is a primary amine. We discovered that changes due to methylation produce striking effects on the average properties of model membranes and further modulate their dynamic behavior. Previous studies have shown the effective area per molecule is less for DPPE than for DPPC, which is due to the smaller size and capacity of phosphatidylethanolamines to hydrogen bond, allowing tighter packing and reduced hydration (37). Close packing of the molecules restricts the lipid cross-sectional area available to the acyl groups and is compensated for by greater chain travel along the director. In consequence, stretching of the chains leads to an increased DPPE bilayer thickness versus DPPC membranes. Further dynamic effects have been uncovered in this work that we attribute to greater bending rigidity of PE-containing bilayers, with implications for lipid nanostructures and lipid modulation of protein function (89).
On account of the different behavior of the polar headgroups (4,22,31), modulation of membrane properties is possible by mixing lipids of different molecular areas or bilayer thickness (89). In the single-phase region of mixtures of phospholipids such as those found in biomembranes, the average properties depend on both the polar headgroups and the fatty acyl composition (7). Inclusion of phosphatidylethanolamine in the bilayer has the effect of condensing the membrane, making it thicker in the ld phase. Methylation of the headgroups, as in the case of PC versus PE, weakens the intermolecular interactions, leading to greater area per lipid, smaller bilayer thickness, and a lower order-disorder phase transition temperature. Headgroup methylation also decreases the absolute spontaneous curvature of the individual monolayer leaflets of the bilayer, favoring lamellar nanostructures and increasing the window of stability between the so (gel) and nonlamellar (HII) phases. Altering the lipid composition thus gives an important means for modulating structural or material properties of cellular membranes in relation to protein activity.
Structure and mobility of phospholipids in the solid-ordered phase are revealed by NMR spectroscopy
Additionally, the low-temperature behavior of DPPE-d62 varies from the DPPC-d62 bilayer and other diacylphosphatidylcholines (72) due to the different headgroup interactions. In the so phase of DPPE-d62 the solid-state 2H NMR spectra have the same approximate shape as expected for all-trans, axially rotating acyl chains. Nonetheless, the observed spectral moments are smaller than expected. The first and second moments are calculated to be 1.5 × 105 s−1 and 2.9 × 1010 s−2, respectively, for the case of rotating polymethylene chains with rotating methyl groups (55). These values are higher than those in Fig. 3, indicating that, even in the so phase, there is appreciable disorder. By contrast, for the DPPC-d62 bilayer, a broad distribution of methylene splittings is evident, which lacks the well-defined value of ±23 kHz (Fig. 2 C). When the temperature is decreased to below 30°C, the spectra for DPPE-d62 change and they lose the distinct splitting at ±23 kHz. The solid-state 2H NMR spectra and the moments of both the DPPE-d62 and DPPC-d62 multilamellar dispersions (Figs. 2 B and C and 3) then become rather similar.
Compared with phosphatidylethanolamines, the diacyl phosphatidylcholines have a broader distribution of quadrupolar splittings along the chains, because they are not fully in the all-trans state. In addition, rotation about the long molecular axis could be in the intermediate exchange region on the NMR time scale for the diacylphosphatidylcholines. The lineshape simulations indicate that this hypothesis can account for much of the differences (56). Current lineshape simulations of DPPE-d62 and DPPC-d62 (Fig. 2 A–C) indicate that variations in the axial diffusion rates can produce changes in the solid-state 2H NMR spectra. At 40°C, the DPPE-d62 molecules undergo axial diffusion at approximately 6.6 × 106 rad s−1, which is rapid on the 2H NMR time scale. When the temperature is lowered, this rate is diminished, and at 29°C it is slow enough to affect the solid-state 2H NMR lineshape (2.9 × 106 rad s−1). This rate is comparable with DPPC-d62 at the same temperature, where the two bilayer systems yield similar spectra. On the other hand, DPPC-d62 exhibits such spectra at all temperatures below the chain melting phase transition. The higher order-disorder phase transition temperatures for phosphatidylethanolamines can be understood in terms of hydrogen bonding of the polar headgroups (90). Conversely, the lack of a pretransition in DPPE-d62 is likely also related to the size of the headgroups. For DPPC-d62, by contrast, once the chains are in the all-trans conformation, they must tilt to compensate for the cross-sectional area, as they are not large enough to fill the space beneath the phosphocholine headgroups. Because the phosphoethanolamine headgroups are much smaller, the chains do not need to tilt once they become all-trans (81), and hence no pretransition is observed.
Segmental order parameters describe average phospholipid structure in the liquid-disordered phase
Following this logic, the effective size of the polar headgroups also affects the average properties of the acyl chains in the ld (or Lα) phase. The different order profiles of DPPE-d62 and DPPC-d62 must be related to methylation of the polar headgroups because the acyl chains are identical (Fig. 5). As mentioned above, the most obvious distinction is the headgroup size and capacity for hydrogen bonding (91,92). Both X-ray diffraction and solid-state 2H NMR spectroscopy indicate the phosphatidylethanolamines occupy a smaller area per lipid at the aqueous interface compared with phosphatidylcholines. Based on our solid-state 2H NMR results, the smaller interfacial area per molecule for DPPE-d62 is likely associated with the larger order parameters compared with the DPPC-d62 bilayer (93). The lack of methyl groups and hydrogen bonding of the primary amine yields an overall reduction in the average area per molecule. Hence, for DPPE-d62, the chains must become more ordered, i.e., stretched along the bilayer normal, to conserve the density at that of liquid hydrocarbon.
An added corollary is that the order parameters are directly relevant to the balance of attractive and repulsive forces acting upon the polar headgroups and the acyl chains. One possibility is for the lipid volume to take an average shape that optimally packs into a particular nanostructure (94). For example, lipids in a planar bilayer approximate the shape of a cylinder or rectangular parallelepiped, in which the area at the lipid-water interface is the same as the average cross-sectional area of the chains. If the area at the lipid-water interface is smaller (i.e., due to substituting PE for PC), then the cylinder must be longer, because the chain volume remains constant. Longer acyl chains mean fewer gauche isomers and higher order parameters, as seen experimentally. The average shape concept is useful in interpreting the solid-state 2H NMR order profiles and when comparing the order parameters of lipid phases (28). The PE headgroups form a smaller cross-sectional area relative to the acyl chain, resulting in an inverted cone shape for lipid molecules and thus having negative spontaneous curvature, whereas PC headgroup lipids form a cylindrical molecular shape, leading to zero spontaneous curvature (7,95). The elastic deformation is formulated in terms of the dynamic bending rigidity using NMR relaxation methods. Accordingly, an explanation in terms of spontaneous curvature may be informative (95). Because of the small size of the phosphoethanolamine headgroups, they favor a reduced cross-sectional area versus the chains. The spontaneous (intrinsic) curvature or bending moment of an individual lipid monolayer (leaflet) toward water is described by the flexible surface model (FSM) (7). It corresponds to bending of a neutral plane, where the area per lipid is constant, so that the deformation energy is described by the mean-curvature modulus (bending rigidity). Monolayer bending is counteracted by the stretching energy of the chains in the two opposed monolayers, as revealed by the 2H NMR order profiles. In the lamellar phase, the curvature is zero, leading to frustration of the spontaneous curvature (88,95). For either picture, the smaller interfacial area and reduced hydration of PE headgroups lead to greater lipid strain deformation in the lamellar phase that can affect membrane protein structure and activity.
Next, we can ask what happens when lipids with different headgroup sizes, i.e., DPPE and DPPC, are mixed in the ld state. In such cases, the segmental order parameters of the acyl chains are intermediate between those of the pure components and are independent of the identity of the headgroup in the phospholipid mixtures (Fig. 7 A). Let us now come back to the example of mixtures of lipids with a similar chemical composition. For example, mixtures of DPPC and 1-palmitoyl-sn-glycero-3-phosphocholine (PalPC) form bilayers whose properties are averaged over all the lipids in the system (56,96). What is striking is that this average can be detected no matter which component is deuterated. Similarly, for the DPPE/DPPC mixtures, the fact that in the ld phase the lipid order parameters are identical indicates that the headgroup to which the deuterated acyl chains are attached is not the determining factor. The headgroups form the surface of the lipid nanostructure and govern the cross-sectional area available to the acyl chains in the ld phase. As a result, changing the amounts of different components has an effect that is averaged over the entire membrane.
Solid-state NMR relaxation experimentally investigates the molecular dynamics of phospholipid membranes
Up to this point we have mainly considered the differences in equilibrium or average properties of the DPPE-d62 and DPPC-d62 bilayers. However, it is likely that variations in dynamic properties may be equally significant, as quantified by the rates and amplitudes of the structural fluctuations (23). We have already pointed out how the thermal motions of the lipids are subject to the same forces and potentials that govern their equilibrium properties (85). According to the fluctuation-dissipation theorem, if an object is deformed (in our case, a lipid membrane), then kinetic energy is dissipated as heat. In the reverse process, the spontaneous thermal fluctuations around the mean structure (measured via nuclear spin relaxation) depend on the same material constants, e.g., the bending rigidity (κ) (85,97,98). In solid-state 2H NMR spectroscopy, the fluctuations of the EFG tensor are associated with the C−2H bond direction to the main magnetic field and are the origin of the relaxation. Motions of the C−2H bond segment, e.g., due to trans–gauche isomerizations, cause the orientation of the molecule-fixed tensor to fluctuate, which is monitored in relaxometry experiments (99). The various motions correspond to local isomerizations of the lipids, together with collective segmental or molecular motions related to the intermolecular forces. Each of the motional types with different mean-square amplitudes has a function Gm(τ) that describes how the orientation of the EFG tensor at time t is correlated to its orientation at a time t + τ later. For a stationary Markov process, the spectral densities Jm(mω0) of the motions (power spectra) are related to the nuclear spin relaxation rates (Eqs. 12a and 12b) and are given by Fourier transformation of the correlation functions.
If only one relaxation rate is measured, then clearly the irreducible spectral densities of the bilayer motions cannot be separated. However, by measuring both the R1Z and R1Q rates, the spectral densities J1(ω0) and J2(2ω0) can be obtained as model-free spectroscopic observables (Fig. 6 B). Because the local motions are very rapid—the bilayer microviscosity is on the order of liquid hydrocarbon (62,85,100)—collective motions of the lipid molecules can be dominant; e.g., due to bilayer twist, splay, and bend deformations (85,101). Rapid segmental motions can pre-average the static EFG tensor, leading to a residual or effective tensor, which is modulated by the molecular or collective bilayer disturbances that cause the relaxation. In terms of the fluctuation-dissipation theorem (85), the thermal bilayer dynamics correspond to its deformation under an applied external force. Pre-averaging of the EFG tensor by rapid localized motions would mean the collective order-director fluctuations (ODF) lead to smaller relaxation rates and spectral densities as a result (83). This explanation can account for the decrease in relaxation rates as the temperature is raised, or toward the ends of the acyl chains. Still, it would then be expected that the relaxation rates for DPPE-d62 exceed those of the DPPC-d62 bilayer if the viscoelasticity is the same, due to the increased order parameters, which is not seen (Fig. 6). Consequently, the segmental order parameter by itself does not explain the differences in relaxation rates; additional factors such as the bending rigidity (κ) must be considered (102).
Collective dynamics and emergent elasticity of lipid membranes are revealed by nuclear spin relaxation
Experimentally, the relaxation rate profiles as a function of chain position in the ld phase are related to the corresponding SCD order parameter profiles. Because the nuclear spin relaxation rates and the corresponding spectral densities depend on both the rates and amplitudes of the lipid motions, the relaxation profile is a functional of the order profile; often, there is a model-free, square-law dependence of the relaxation profile on the order profile (Fig. 7 C). How can we explain these striking differences in relaxation rates of the acyl chains, which are due to the lipid polar headgroups? One explanation is that fast segmental fluctuations of the lipids set up an order profile that is pre-averaged due to localized trans–gauche isomerizations. The local order profile is then further averaged by collective bilayer fluctuations of the lipids to give the observed order profile (85). As noted above, the static coupling constant (due to the electric quadrupolar interaction in 2H NMR spectroscopy) is pre-averaged by the local fluctuations, giving a residual coupling constant that can be further modulated by collective bilayer disturbances due to ODF of the lipids. Because the relaxation rates and spectral densities depend on the spectral power of the fluctuations—i.e., the square of the coupling constant (Fermi’s golden rule) (103)—such a square-law relation is immediately explained. Collective fluctuations of the lipids are then described within a continuum approximation in terms of material constants for the twist, splay, and bend fluctuations of the bilayer itself. Within a single elastic constant approximation, the coefficient of the collective viscoelastic deformations is related to the Helfrich bending rigidity (κ) (104). In this way, a connection is found between the spontaneous thermal fluctuation of the bilayer detected at an atomistic level and the material elastic constants for deformation away from equilibrium (85,105).
For membrane lipids, we can formulate the spin-lattice relaxation as the result of the composite fast and slow motions (85,105), which leads to
| (13) |
and, similarly,
| (14) |
In the above formulas, the relaxation rates are assumed to be motionally averaged by lateral diffusion of the lipids over the curved bilayer surfaces (53). The square-law dependence of the and relaxation rate profiles on the corresponding order parameter profile in terms of the acyl carbon index (i) is a signature of collective slow motions of the lipids (83,85). In terms of quantum mechanical principles, it is a simple manifestation of Fermi’s golden rule (103). In the first term on the right, is the effective correlation time for fast local motions of the ith segment, and A and C are related to the static coupling constant. The second term entails a distribution of correlation times arising from the collective bilayer modes that undergo viscoelastic relaxation. Slower quasielastic bilayer disturbances in the frequency domain are described by the functions and , where B and D characterize the viscoelasticity of the membrane (97,102,105). A larger bending rigidity gives a smaller slope and vice versa, as universally seen for cholesterol-containing bilayers (21,98,102). An experimental example is shown for the case of the DPPE:DPPC (1:1) lipid mixture (Fig. 7 C), which supports the theoretically predicted (85) square-law dependence of the and relaxation rates on the corresponding order parameters. Remarkably, the structural dynamics are independent of whether DPPE or DPPC is observed as the deuterated lipid species in the binary mixture (Fig. 7 A−C). In mixtures of DPPE and DPPC, the dynamics are the same regardless of the probe lipid headgroup and show intermediate bending rigidities. Average viscoelastic properties of the bilayer are thus detectable with NMR relaxation as they emerge from atomistic interactions.
Until now, the square-law dependence has mainly been shown experimentally for bilayers of disaturated phosphatidylcholines (97,106) in the ld state. The current data extend these results to phosphatidylethanolamines and uncover how the square-law functional relation (Eqs. 13 and 14) corresponds to emergent viscoelastic properties of lipid membranes. Collective lipid motions explain the variation in relaxation between the DPPE-d62 and DPPC-d62 bilayers by differences in their effective viscoelastic constants (85,97,105). In addition, the frequency dependence of the relaxation rates yields an power-law relation (26,62,105) that likewise informs the collective dynamics. Such a relation is also a characteristic signature of the contribution of ODF to the relaxation (107,108), whereby collective modes due to thermal motions lead to fluctuations in the local directors (85). In liquid-crystalline nanomaterials, the director represents the average orientation of the molecules. The local director motions can be visualized as cooperative reorientations of the molecules or their segments relative to the average director (membrane normal), due to quasicoherent, wavelike elastic disturbances that manifest the forces acting on the lipid molecules (62,85).
Our investigations are summarized (Fig. 8) to reveal how the experimental R1Z rate profiles are functionals of the order parameter |SCD| profiles. The data support the square-law relation first described (85) for the DPPC bilayer (Fig. 8 A), and moreover extend the approach to phosphatidylethanolamine systems (Fig. 8 B and C). Comparison of the plots at various temperatures for DPPE-d62 and DPPC-d62 bilayers shows that, in the former case, the ODF contribution is strikingly reduced. According to our picture, the diminished slope for DPPE indicates a higher bending rigidity than for the DPPC bilayer, which emerges at the local atomistic level. Our findings are consistent with the structural parameters calculated using the mean-torque model (Fig. 8 A, inset). The change in collective motions for phosphatidylethanolamine is explained by closer packing of the polar headgroups., whereas the square-law plots for DPPC/DPPE mixtures (Fig. 8 B) fall in between those for the individual lipids (Fig. 8 A and C). As a general result, the square-law dependencies of the relaxation rates for lipid mixtures show similar contributions independent of the labeled molecule. This finding points to averaging of the viscoelastic properties just as for the structural parameters. The combined relaxation rate and order parameter measurements report on viscoelastic properties of the bilayer as they emerge from the atomistic level interactions and are uniquely detected with solid-state NMR spectroscopy.
Figure 8.
Summary of influences of polar headgroups on solid-state 2H NMR spin-lattice relaxation rates and order parameters for multilamellar lipids in the ld phase. Square-law plots of versus are shown for (A) DPPC-d62 bilayers at various temperatures, (B) DPPE-d62:DPPC (1:1) and DPPC-d62/DPPE (1:1) lipid mixtures at 65°C, and (C) DPPE-d62 membranes at different temperatures in the ld phase (Lα phase) (data included from Ref. (97)). Inset shows mean-torque model-derived structural parameters, bilayer steric thickness (solid line), and average interfacial area per lipid (dotted line) at various temperatures. Note that the square-law slopes for the DPPE:DPPC mixtures are independent of the deuterated molecule and have an intermediate value relative to the component lipids. The estimated error in the relaxation rates is ±5% and the error in the order parameters is ±0.001. To see this figure in color, go online.
Implications for lipid biophysics
Solid-state 2H NMR spectroscopy provides exceptional insight into how the phospholipid headgroups control the emergent structural and material properties of biomembranes. Experimentally, the approach is highly synergistic with X-ray (36,107,109,110) and neutron (21,111,112) diffraction studies of symmetric and asymmetric lipid membranes (34). Our investigations highlight how the smaller effective size and hydrogen-bonding capacity of the phosphoethanolamine headgroups affect the acyl chain packing and dynamics compared with methylated phosphocholine substituents. In the so or gel phases, there is significant mobility of the lipids. Collective lipid dynamics at mesoscopic length scales in the ld phase further inform the emergence of soft-material properties, such as the bending rigidity (modulus) (110,113, 114, 115, 116). Close packing of the phosphatidylethanolamine molecules restricts the cross-sectional area available to the acyl chains, yielding higher segmental order parameters and greater bending rigidity versus phosphatidylcholine lipids, with implications for proteolipid coupling mechanisms (7). Phosphoethanolamine lipids enable membrane integrity and may also underlie the different interactions of antimicrobial peptides with gram-positive bacterial membranes versus eukaryotic membranes containing cholesterol (117,118). Our findings contribute to understanding the diversity of lipids in biological membranes within the broader context of membrane protein structure and activity, where elastic deformation and softness of the bilayer have key roles (7,89). In lipid mixtures, the properties of the components are summed to modulate average membrane behavior by changing the composition. An intriguing question is whether the asymmetric lipid distribution in cellular membranes versus symmetric model bilayers affects membrane protein activity or properties such as the bending rigidity. Establishing how the lipid elastic deformation corresponds to the length scales probed by various experimental methods remains as an additional challenging topic of great biophysical significance.
Author contributions
M.F.B. developed the initial concept. R.L.T., T.M.A., and T.P.T performed the experiments. T.R.M., R.L.T., T.M.A., and T.P.T carried out the analysis. T.R.M. and M.F.B. wrote the manuscript with discussion, review, and contributions from all authors.
Acknowledgments
We thank H. Petrache, J. Kinnun, K. Mallikarjuniah, and A. Struts for discussions, and C. Job for expert electronics assistance.
This research was supported by NIH Postdoctoral Fellowship EY06346 (T.M.A.), NIH grants EY012049 and EY02604 (M.F.B.), and NSF awards CHE 1904125 and MCB 11817862 (M.F.B.).
Declaration of interests
The authors declare no competing interests.
Editor: Timothy Cross.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.09.005.
Supporting material
References
- 1.Ingólfsson H.I., Melo M.N., et al. Marrink S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 2.Sezgin E., Levental I., et al. Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorent J.H., Levental K.R., et al. Levental I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020;16:644–652. doi: 10.1038/s41589-020-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullis P.R., Hope M.J., Tilcock C.P. Lipid polymorphism and the roles of lipids in membranes. Chem. Phys. Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- 5.Phillips R., Ursell T., et al. Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauwers E., Goodchild R., Verstreken P. Membrane lipids in presynaptic function and disease. Neuron. 2016;90:11–25. doi: 10.1016/j.neuron.2016.02.033. [DOI] [PubMed] [Google Scholar]
- 7.Brown M.F. Soft matter in lipid–protein interactions. Annu. Rev. Biophys. 2017;46:379–410. doi: 10.1146/annurev-biophys-070816-033843. [DOI] [PubMed] [Google Scholar]
- 8.Gupta K., Donlan J.A.C., et al. Robinson C.V. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulino J., Pang X., et al. Cross T.A. Influenza A M2 channel clustering at high protein/lipid ratios: viral budding implications. Biophys. J. 2019;116:1075–1084. doi: 10.1016/j.bpj.2019.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown M.F., Seelig J. Ion-induced changes in head group conformation of lecithin bilayers. Nature. 1977;269:721–723. [Google Scholar]
- 11.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soubias O., Teague W.E., Jr., et al. Hines K.G. Contribution of membrane elastic energy to rhodopsin function. Biophys. J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozelli J.C., Jr., Epand R.M. Determinants of lipids acyl chain specificity: a tale of two enzymes. Biophys. Chem. 2020;265:106431. doi: 10.1016/j.bpc.2020.106431. [DOI] [PubMed] [Google Scholar]
- 14.Newton A.C. Interactions of proteins with lipid headgroups: lessons from protein kinase C. Annu. Rev. Biophys. Biomol. Struct. 1993;22:1–25. doi: 10.1146/annurev.bb.22.060193.000245. [DOI] [PubMed] [Google Scholar]
- 15.Laganowsky A., Reading E., et al. Robinson C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cady S.D., Schmidt-Rohr K., et al. Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawaliby R., Trubbia C., et al. Govaerts C. Allosteric regulation of G protein–coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baccouch R., Rascol E., et al. Alves I.D. The role of the lipid environment in the activity of G protein coupled receptors. Biophys. Chem. 2022;285:106794. doi: 10.1016/j.bpc.2022.106794. [DOI] [PubMed] [Google Scholar]
- 19.Bozelli J.C., Aulakh S.S., Epand R.M. Membrane shape as determinant of protein properties. Biophys. Chem. 2021;273:106587. doi: 10.1016/j.bpc.2021.106587. [DOI] [PubMed] [Google Scholar]
- 20.Leftin A., Molugu T.R., et al. Brown M.F. Area per lipid and cholesterol interactions in membranes by separated local-field 13C NMR spectroscopy. Biophys. J. 2014;107:2274–2286. doi: 10.1016/j.bpj.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty S., Doktorova M., et al. Ashkar R. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. USA. 2020;117:21896–21905. doi: 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold K., Lösche A., Gawrisch K. 31P-NMR investigations of phase separation in phosphatidylcholine/phosphatidylethanolamine mixtures. Biochim. Biophys. Acta. 1981;645:143–148. doi: 10.1016/0005-2736(81)90522-8. [DOI] [PubMed] [Google Scholar]
- 23.Brown M.F., Seelig J., Häberlen U. Structural dynamics in phospholipid bilayers from deuterium spin-lattice relaxation time measurements. J. Chem. Phys. 1979;70:5045–5053. [Google Scholar]
- 24.Davis J.H., Bloom M., et al. Smith I.C. The temperature dependence of molecular order and the influence of cholesterol in Acholeplasma laidlawii membranes. Biochim. Biophys. Acta. 1980;597:477–491. doi: 10.1016/0005-2736(80)90221-7. [DOI] [PubMed] [Google Scholar]
- 25.Leftin A., Xu X., Brown M.F. Phospholipid bilayer membranes: deuterium and carbon-13 NMR spectroscopy. eMagRes. 2014;3:199–214. [Google Scholar]
- 26.Xu X., Struts A.V., Brown M.F. Generalized model-free analysis of nuclear spin relaxation experiments. eMagRes. 2014;3:275–286. [Google Scholar]
- 27.Molugu T.R., Lee S., Brown M.F. Concepts and methods of solid-state NMR spectroscopy applied to biomembranes. Chem. Rev. 2017;117:12087–12132. doi: 10.1021/acs.chemrev.6b00619. [DOI] [PubMed] [Google Scholar]
- 28.Thurmond R.L., Lindblom G., Brown M.F. Curvature, order, and dynamics of lipid hexagonal phases studied by deuterium NMR spectroscopy. Biochemistry. 1993;32:5394–5410. doi: 10.1021/bi00071a015. [DOI] [PubMed] [Google Scholar]
- 29.Molugu T.R., Brown M.F. Cholesterol-induced suppression of membrane elastic fluctuations at the atomistic level. Chem. Phys. Lipids. 2016;199:39–51. doi: 10.1016/j.chemphyslip.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molugu T.R., Xu X., et al. Brown M.F. In: Modern Magnetic Resonance. Webb G.A., editor. Springer International Publishing; Cham: 2018. Solid-state 2H NMR studies of water-mediated lipid membrane deformation; pp. 1–27. [Google Scholar]
- 31.Lafleur M., Bloom M., et al. Cullis P.R. Correlation between lipid plane curvature and lipid chain order. Biophys. J. 1996;70:2747–2757. doi: 10.1016/S0006-3495(96)79844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toppozini L., Meinhardt S., et al. Rheinstädter M.C. Structure of cholesterol in lipid rafts. Phys. Rev. Lett. 2014;113:228101–228105. doi: 10.1103/PhysRevLett.113.228101. [DOI] [PubMed] [Google Scholar]
- 33.Kinnun J.J., Mallikarjunaiah K.J., et al. Brown M.F. Elastic deformation and area per lipid of membranes: atomistic view from solid-state deuterium NMR spectroscopy. Biochim. Biophys. Acta. 2015;1848:246–259. doi: 10.1016/j.bbamem.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heberle F.A., Marquardt D., et al. Pabst G. Subnanometer structure of an asymmetric model membrane: interleaflet coupling influences domain properties. Langmuir. 2016;32:5195–5200. doi: 10.1021/acs.langmuir.5b04562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veatch S.L., Soubias O., et al. Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrache H.I., Dodd S.W., Brown M.F. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectrscopy. Biophys. J. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor R.W., Venable R.M., Feller S.E. Lipid bilayers, NMR relaxation, and computer simulations. Acc. Chem. Res. 2002;35:438–446. doi: 10.1021/ar0100529. [DOI] [PubMed] [Google Scholar]
- 39.Venable R.M., Brown F.L.H., Pastor R.W. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem. Phys. Lipids. 2015;192:60–74. doi: 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrink S.J., Corradi V., et al. Sansom M.S.P. Computational modeling of realistic cell membranes. Chem. Rev. 2019;119:6184–6226. doi: 10.1021/acs.chemrev.8b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yesylevskyy S.O., Rivel T., Ramseyer C. The influence of curvature on the properties of the plasma membrane. Insights from atomistic molecular dynamics simulations. Sci. Rep. 2017;7:16078. doi: 10.1038/s41598-017-16450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodt A.J., Pastor R.W., Lyman E. Hexagonal substructure and hydrogen bonding in liquid-ordered phases containing palmitoyl sphingomyelin. Biophys. J. 2015;109:948–955. doi: 10.1016/j.bpj.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mihailescu M., Sorci M., et al. Cotten M.L. Structure and function in antimicrobial piscidins: histidine position, directionality of membrane insertion, and pH-dependent permeabilization. J. Am. Chem. Soc. 2019;141:9837–9853. doi: 10.1021/jacs.9b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber T., Botelho A.V., et al. Brown M.F. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys. J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soubias O., Teague W.E., Jr., et al. Hines K.G. Rhodopsin/lipid hydrophobic matching−rhodopsin oligomerization and function. Biophys. J. 2015;108:1125–1132. doi: 10.1016/j.bpj.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mason J.T., Broccoli A.V., Huang C. A method for the synthesis of isomerically pure saturated mixed-chain phosphatidylcholines. Anal. Biochem. 1981;113:96–101. doi: 10.1016/0003-2697(81)90049-x. [DOI] [PubMed] [Google Scholar]
- 47.Bloom M., Davis J.H., MacKay A.L. Direct determination of the oriented sample NMR spectrum from the powder spectrum for systems with local axial symmetry. Chem. Phys. Lett. 1981;80:198–202. [Google Scholar]
- 48.Sternin E., Bloom M., MacKay A.L. De-Pake-ing of NMR spectra. J. Magn. Reson. 1983;55:274–282. [Google Scholar]
- 49.Greenfield M.S., Ronemus A.D., et al. Raidy T. Deuterium quadrupole-echo NMR spectroscopy. III. Practical aspects of lineshape calculations for multiaxis rotational processes. J. Magn. Reson. 1987;72:89–107. [Google Scholar]
- 50.Vold R.R., Bodenhausen G. Phase-shifted pulse sequence for measurement of spin-lattice relaxation in complex spin systems: an improvement. J. Magn. Reson. 1980;39:363–366. [Google Scholar]
- 51.Wimperis S. Broadband and narrowband composite excitation sequences. J. Magn. Reson. 1990;86:46–59. [Google Scholar]
- 52.Nevzorov A.A., Trouard T.P., Brown M.F. Lipid bilayer dynamics from simultaneous analysis of orientation and frequency dependence of deuterium spin-lattice and quadrupolar order relaxation. Phys. Rev. E. 1998;58:2259–2281. [Google Scholar]
- 53.Brown M.F., Davis J.H. Orientation and frequency dependence of the deuterium spin-lattice relaxation in multilamellar phospholipid dispersions: implications for dynamic models of membrane structure. Chem. Phys. Lett. 1981;79:431–435. [Google Scholar]
- 54.Seelig J., Seelig A. Lipid conformation in model membranes and biological membranes. Q. Rev. Biophys. 1980;13:19–61. doi: 10.1017/s0033583500000305. [DOI] [PubMed] [Google Scholar]
- 55.Davis J.H. Deuterium magnetic resonance study of the gel and liquid crystalline phases of dipalmitoylphosphatidylcholine. Biophys. J. 1979;27:339–358. doi: 10.1016/S0006-3495(79)85222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jansson M., Thurmond R.L., et al. Brown M.F. Deuterium NMR study of intermolecular interactions in lamellar phases containing palmitoyllysophosphatidylcholine. J. Phys. Chem. 1992;96:9532–9544. [Google Scholar]
- 57.Smith A.A., Vogel A., et al. Huster D. A method to construct the dynamic landscape of a bio-membrane with experiment and simulation. Nat. Commun. 2022;13:108. doi: 10.1038/s41467-021-27417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagle J.F. Area/lipid of bilayers from NMR. Biophys. J. 1993;64:1476–1481. doi: 10.1016/S0006-3495(93)81514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber T., Rajamoorthi K., et al. Brown M.F. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by 2H NMR and molecular dynamics simulations. J. Am. Chem. Soc. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 60.Nagle J.F., Wilkinson D.A. Lecithin bilayers. Density measurements and molecular interactions. Biophys. J. 1978;23:159–175. doi: 10.1016/S0006-3495(78)85441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrache H.I., Salmon A., Brown M.F. Structural properties of docosahexaenoyl phospholipid bilayers investigated by solid-state 2H NMR spectroscopy. J. Am. Chem. Soc. 2001;123:12611–12622. doi: 10.1021/ja011745n. [DOI] [PubMed] [Google Scholar]
- 62.Brown M.F., Ribeiro A.A., Williams G.D. New view of lipid bilayer dynamics from 2H and 13C NMR relaxation time measurements. Proc. Natl. Acad. Sci. USA. 1983;80:4325–4329. doi: 10.1073/pnas.80.14.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dill K.A., Flory P.J. Molecular organization in micelles and vesicles. Proc. Natl. Acad. Sci. USA. 1981;78:676–680. doi: 10.1073/pnas.78.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mallikarjunaiah K.J., Kinnun J.J., et al. Brown M.F. Flexible lipid nanomaterials studied by NMR spectroscopy. Phys. Chem. Chem. Phys. 2019;21:18422–18457. doi: 10.1039/c8cp06179c. [DOI] [PubMed] [Google Scholar]
- 65.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown M.F. Deuterium relaxation and molecular dynamics in lipid bilayers. J. Magn. Reson. 1979;35:203–215. [Google Scholar]
- 67.Petrache H.I., Tristram-Nagle S., Nagle J.F. Fluid phase structure of EPC and DMPC bilayers. Chem. Phys. Lipids. 1998;95:83–94. doi: 10.1016/s0009-3084(98)00068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kučerka N., Tristram-Nagle S., Nagle J.F. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 69.Petrache H.I., Feller S.E., Nagle J.F. Determination of component volumes of lipid bilayers from simulations. Biophys. J. 1997;72:2237–2242. doi: 10.1016/S0006-3495(97)78867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huber H. Deuterium quadrupole coupling constants. A theoretical investigation. J. Chem. Phys. 1985;83:4591–4598. [Google Scholar]
- 71.Spiess H.W. In: NMR Basic Principles and Progress. Diehl P., Fluck E., Kosfeld R., editors. Springer-Verlag; Heidelberg: 1978. Rotation of molecules and nuclear spin relaxation; pp. 55–214. [Google Scholar]
- 72.Barry J.A., Trouard T.P., et al. Brown M.F. Low-temperature 2H NMR spectroscopy of phospholipid bilayers containing docosahexaenoyl (22:6ω3) chains. Biochemistry. 1991;30:8386–8394. doi: 10.1021/bi00098a016. [DOI] [PubMed] [Google Scholar]
- 73.Marsh D. CRC Press; Boca Raton: 2013. Handbook of Lipids. [Google Scholar]
- 74.Akabori K., Nagle J.F. Structure of the DMPC lipid bilayer ripple phase. Soft Matter. 2015;11:918–926. doi: 10.1039/c4sm02335h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khakbaz P., Klauda J.B. Investigation of phase transitions of saturated phosphocholine lipid bilayers via molecular dynamics simulations. Biochim. Biophys. Acta. 2018;1860:1489–1501. doi: 10.1016/j.bbamem.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 76.Ruocco M.J., Shipley G. Characterization of the sub-transiton of hydrated dipalmitoylphosphatidylcholine bilayers kinetic hydration and structural study. Biochim. Biophys. Acta. 1982;691:309–320. [Google Scholar]
- 77.Chapman D. Phase transition and fluidity characteristics of lipids and cell membranes. Q. Rev. Biophys. 1975;8:185–235. doi: 10.1017/s0033583500001797. [DOI] [PubMed] [Google Scholar]
- 78.Marsh D., Watts A., Smith I.C. Dynamic structure and phase behavior of dimyristoylphosphatidylethanolamine bilayers studied by deuterium nuclear magnetic resonance. Biochemistry. 1983;22:3023–3026. doi: 10.1021/bi00281a036. [DOI] [PubMed] [Google Scholar]
- 79.Madrid E., Horswell S.L. Effect of headgroup on the physicochemical properties of phospholipid bilayers in electric fields: size matters. Langmuir. 2013;29:1695–1708. doi: 10.1021/la304455d. [DOI] [PubMed] [Google Scholar]
- 80.Lafleur M., Cullis P.R., et al. Bloom M. Comparison of the orientational order of lipid chains in the Lα and HII phases. Biochemistry. 1990;29:8325–8333. doi: 10.1021/bi00488a018. [DOI] [PubMed] [Google Scholar]
- 81.Mendelsohn R., Davies M.A., et al. Dluhy R.A. Quantitative determination of conformational disorder in the acyl chains of phospholipid bilayers by infrared spectroscopy. Biochemistry. 1989;28:8934–8939. doi: 10.1021/bi00448a037. [DOI] [PubMed] [Google Scholar]
- 82.Leftin A., Brown M.F. An NMR database for simulations of membrane dynamics. Biochim. Biophys. Acta. 2011;1808:818–839. doi: 10.1016/j.bbamem.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Williams G.D., Beach J.M., et al. Brown M.F. Dependence of deuterium spin-lattice relaxation rates of multilamellar phospholipid dispersions on orientational order. J. Am. Chem. Soc. 1985;107:6868–6873. [Google Scholar]
- 84.Trouard T.P., Alam T.M., et al. Brown M.F. Angular anisotropy of 2H NMR spectral densities in phospholipid bilayers containing cholesterol. Chem. Phys. Lett. 1992;189:67–75. [Google Scholar]
- 85.Brown M.F. Theory of spin-lattice relaxation in lipid bilayers and biological membranes. 2H and 14N quadrupolar relaxation. J. Chem. Phys. 1982;77:1576–1599. [Google Scholar]
- 86.Blume A., Wittebort R.J., et al. Griffin R.G. Phase equilibria, molecular conformation, and dynamics in phosphatidylcholine/phosphatidylethanolamine bilayers. Biochemistry. 1982;21:6243–6253. doi: 10.1021/bi00267a032. [DOI] [PubMed] [Google Scholar]
- 87.Fried S.D.E., Lewis J.W., et al. Brown M.F. Membrane curvature revisited the archetype of rhodopsin studied by time-resolved electronic spectroscopy. Biophys. J. 2021;120:440–452. doi: 10.1016/j.bpj.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 89.Brown M.F. In: Current Topics in Membranes. Epand R.M., editor. Academic Press; San Diego: 1997. Influence of nonlamellar-forming lipids on rhodopsin; pp. 285–356. [Google Scholar]
- 90.Nagle J.F. Theory of the main lipid phase transition. Annu. Rev. Phys. Chem. 1980;31:157–196. [Google Scholar]
- 91.Wilkinson D.A., Nagle J.F. Dilatometry and calorimetry of saturated phosphatidylethanolamine dispersions. Biochemistry. 1981;20:187–192. doi: 10.1021/bi00504a031. [DOI] [PubMed] [Google Scholar]
- 92.Nagle J.F., Wiener M.C. Structure of fully hydrated bilayer dispersions. Biochim. Biophys. Acta. 1988;942:1–10. doi: 10.1016/0005-2736(88)90268-4. [DOI] [PubMed] [Google Scholar]
- 93.Meraldi J.-P., Schlitter J. A statistical mechanical treatment of fatty acyl chain order in phospholipid bilayers and correlation with experimental data. Biochim. Biophys. Acta. 1981;645:183–192. doi: 10.1016/0005-2736(81)90189-9. [DOI] [PubMed] [Google Scholar]
- 94.Israelachvili J.N., Mitchell D.J., Ninham B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc., Faraday Trans. 2. 1976;72:1525–1568. [Google Scholar]
- 95.Brown M.F. Curvature forces in membrane lipid-protein interactions. Biochemistry. 2012;51:9782–9795. doi: 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jansson M., Thurmond R.L., et al. Brown M.F. Magnetic alignment and orientational order of dipalmitoylphosphatidylcholine bilayers containing palmitoyllyso-phosphatidylcholine. Chem. Phys. Lipids. 1990;54:157–170. doi: 10.1016/0009-3084(90)90009-g. [DOI] [PubMed] [Google Scholar]
- 97.Brown M.F., Thurmond R.L., et al. Beyer K. Composite membrane deformation on the mesoscopic length scale. Phys. Rev. E. 2001;64:010901. doi: 10.1103/PhysRevE.64.010901. [DOI] [PubMed] [Google Scholar]
- 98.Martinez G.V., Dykstra E.M., et al. Brown M.F. NMR elastometry of fluid membranes in the mesoscopic regime. Phys. Rev. E. 2002;66:050902. doi: 10.1103/PhysRevE.66.050902. [DOI] [PubMed] [Google Scholar]
- 99.Brown M.F., Chan S.I. eMagRes. John Wiley & Sons, Ltd; 2007. Bilayer membranes: deuterium and carbon-13 NMR; pp. 1–15. [Google Scholar]
- 100.Venable R.M., Zhang Y., et al. Pastor R.W. Molecular dynamics simulations of a lipid bilayer and of hexadecane: an investigation of membrane fluidity. Science. 1993;262:223–226. doi: 10.1126/science.8211140. [DOI] [PubMed] [Google Scholar]
- 101.Bonmatin J.-M., Smith I.C.P., et al. Siminovitch D.J. Use of a comprehensive approach to molecular dynamics in ordered lipid systems: cholesterol reorientation in ordered lipid bilayers. A 2H NMR relaxation case study. J. Am. Chem. Soc. 1990;112:1697–1704. [Google Scholar]
- 102.Ashkar R., Doktorova M., et al. Brown M.F. Reply to Nagle et al.: the universal stiffening effects of cholesterol on lipid membranes. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2102845118. e2102845118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cohen-Tannoudji C. Herman; Paris: 1978. Quantum Mechanics. [Google Scholar]
- 104.Helfrich W., Servuss R.-M. Undulations, steric interaction and cohesion of fluid membranes. Il Nuovo Cimento D. 1984;3:137–151. [Google Scholar]
- 105.Nevzorov A.A., Brown M.F. Dynamics of lipid bilayers from comparative analysis of 2H and 13C nuclear magnetic resonance relaxation data as a function of frequency and temperature. J. Chem. Phys. 1997;107:10288–10310. [Google Scholar]
- 106.Brown M.F., Thurmond R.L., et al. Beyer K. Elastic deformation of membrane bilayers probed by deuterium NMR relaxation. J. Am. Chem. Soc. 2002;124:8471–8484. doi: 10.1021/ja012660p. [DOI] [PubMed] [Google Scholar]
- 107.Salditt T., Vogel M., Fenzl W. Thermal fluctuations and positional correlations in oriented lipid membranes. Phys. Rev. Lett. 2003;90:178101. doi: 10.1103/PhysRevLett.90.178101. [DOI] [PubMed] [Google Scholar]
- 108.Dong R.Y. In: Annual Reports on NMR Spectroscopy. Webb G.A., editor. Academic Press; 2016. Recent NMR studies of thermotropic liquid crystals; pp. 41–174. [Google Scholar]
- 109.Pabst G., Rappolt M., et al. Laggner P. Structural information from multilamellar liposomes at full hydration: full q-range fitting with high quality x-ray data. Phys. Rev. E. 2000;62:4000–4009. doi: 10.1103/physreve.62.4000. [DOI] [PubMed] [Google Scholar]
- 110.Pan J., Mills T.T., et al. Nagle J.F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. Lett. 2008;100:198103. doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Endress E., Heller H., et al. Bayerl T.M. Anisotropic motion and molecular dynamics of cholesterol, lanosterol, and ergosterol in lecithin bilayers studied by quasi-elastic neutron scattering. Biochemistry. 2002;41:13078–13086. doi: 10.1021/bi0201670. [DOI] [PubMed] [Google Scholar]
- 112.Kučerka N., Nieh M.-P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 113.Sorre B., Callan-Jones A., et al. Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tian A., Capraro B.R., et al. Baumgart T. Bending stiffness depends on curvature of ternary lipid mixture tubular membranes. Biophys. J. 2009;97:1636–1646. doi: 10.1016/j.bpj.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gracià R.S., Bezlyepkina N., et al. Dimova R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter. 2010;6:1472–1482. [Google Scholar]
- 116.Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interface Sci. 2014;208:225–234. doi: 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 117.Doole F.T., Kit Chan C., et al. Brown M.F. Rivalry of cholesterol and antimicrobial peptides as seen by molecular simulations and NMR spectroscopy. Biophys. J. 2022;121:161a–162a. [Google Scholar]
- 118.Doole F.T., Kumarage T., et al. Brown M.F. Cholesterol stiffening of model mammalian and microbial membranes. Biophys. J. 2022;121:34a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.