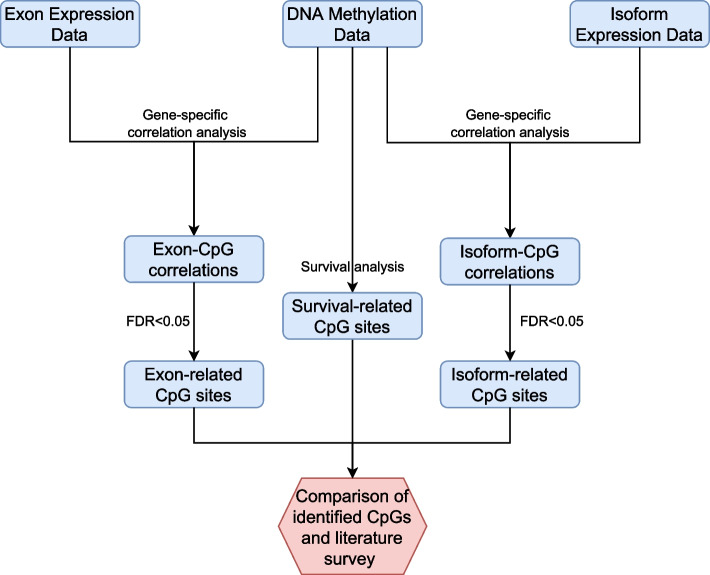
Fig. 1.
Flowchart of data analysis used in this study. First, we selected gene-specific samples that highly expressed the gene for each of the genes we analysed, and then we performed correlation analysis between DNA methylation and exon/isoform expression based on gene-specific samples. A threshold of FDR<0.05 was required to detect significant correlations. In addition, we performed survival analysis for each CpG sites and identified a list of CpG sites that are predictive of patients’ survival outcome. Finally, a literature survey was conducted to search for existing evidences of identified correlations