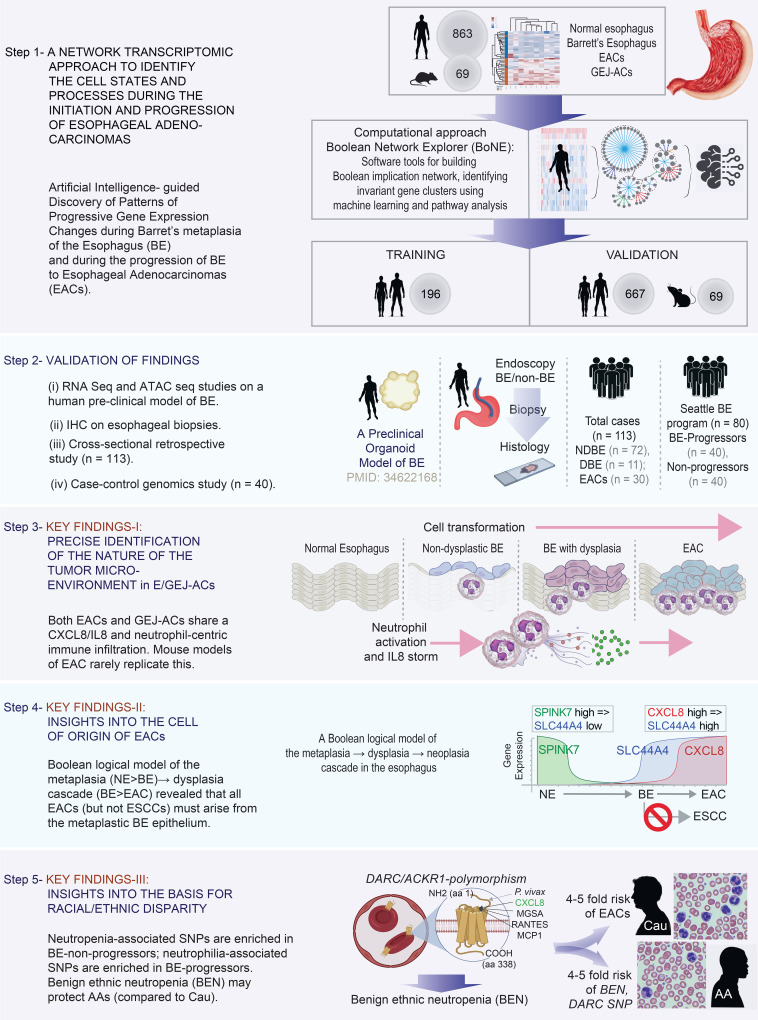
Figure 1. Study design.
Step 1: Numerous transcriptomic data sets from both human (n = 863) and mouse (n = 69) samples were mined to build a validated Boolean implication network–based computational model of disease continuum states during the metaplastic→dysplastic→neoplastic cascade in the squamous epithelial lining of the esophagus. Gene signatures derived from the network-based model are first prioritized by machine-learning approaches and used subsequently to discover cell types and cellular states that fuel the cascade. Step 2: Network predictions were validated in 4 different models and approaches. Steps 3–5: Summary of key conclusions. ATAC seq, assay for transposase-accessible chromatin followed by high-throughput sequencing; EAC/GEJ-AC, esophageal adenocarcinoma/gastroesophageal carcinoma.