Figure 6. ATP13A1 protects misoriented proteins from ERAD.
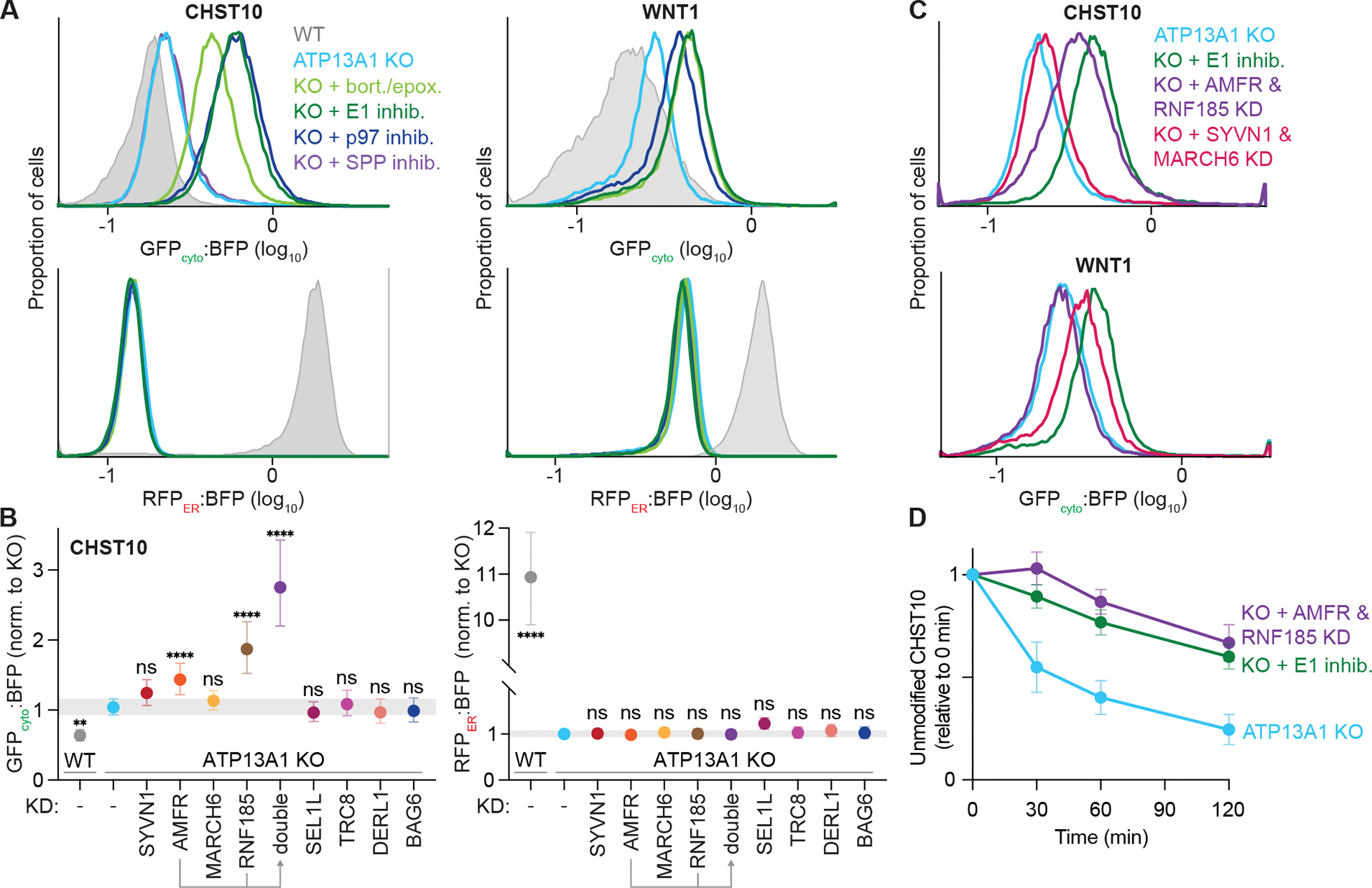
(A) Inhibiting the ubiquitin-proteasome system stabilizes misoriented proteins. GFPcyto:BFP (top) and RFPER:BFP (bottom) ratios of the CHST10 type II (left) or WNT1 signal sequence (right) topology reporter in wildtype (WT; gray) or ATP13A1 knockout (KO; light blue) cells treated without or with inhibitors (inhib.) of SPP (purple), proteasomal activity (bort./epox.; light green), the E1 ubiquitin activating enzyme (1 μM MLN7243; dark green), or the p97 AAA-ATPase (1 μM CB-5083; dark blue).
(B) Distinct ubiquitin ligases mediate misoriented CHST10 ERAD. GFPcyto:BFP (left) and RFPER:BFP (right) ratios of the CHST10 topology reporter in WT (gray) ATP13A1 KO cells treated with siRNAs to knock down (KD) the indicated factors. Shown are median values and interquartile range of at least two biological replicates of n≥5,000 each.
(C) Specific but redundant ubiquitin ligases mediate misoriented protein ERAD. GFPcyto:BFP ratios of the CHST10 (top) or WNT1 (bottom) topology reporter in ATP13A1 KO cells treated without (light blue) or with E1 inhibitor (dark green), or with siRNAs to knock down either AMFR and RNF185 (purple) or SYVN1 and MARCH6 (pink).
(D) Timecourse of misoriented CHST10 ERAD. Translation shut-off reactions of ATP13A1 KO cells expressing the CHST10 topology reporter treated without (light blue) or with siRNAs against AMFR and RNF185 (purple) or E1 inhibitor (dark green). Samples taken at the indicated timepoints were analyzed by immunoblotting for unmodified CHST10, normalized to time = 0 min, and the mean +/− sem for 3 replicates plotted.
See also Figure S7.
