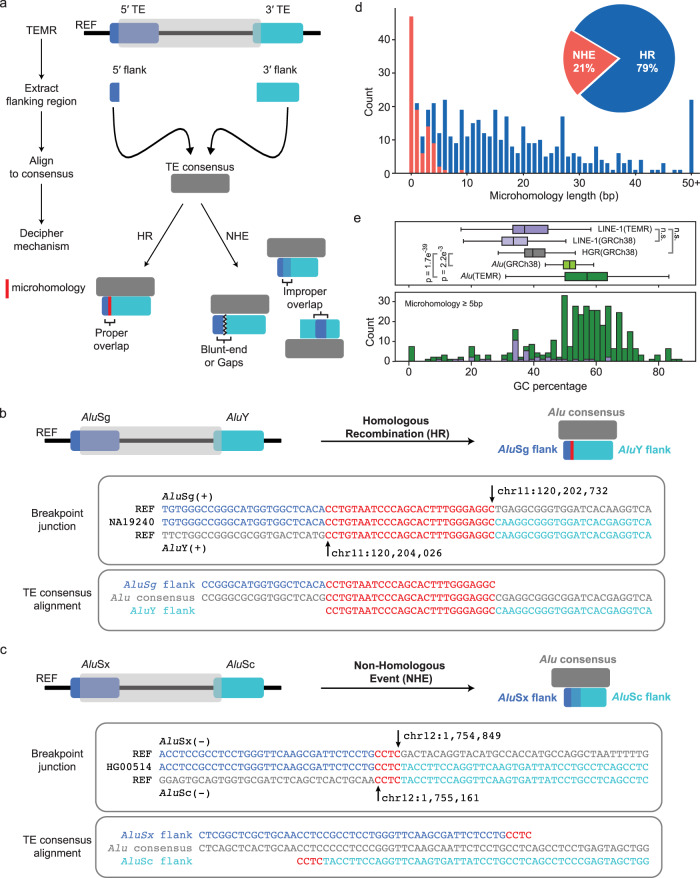
Fig. 2. Identifying mechanistic signatures of TEMRs.
a TEMR events are classified by breakpoint characteristics guided by TE consensus sequences. For homologous breakpoints, the chimeric TE resulting from the TEMR event must reconstruct a full TE with microhomology at the breakpoint. HR, homologous recombination; NHE, non-homologous repair; TE, transposable element; SV, structural variant; MEI, mobile element insertion, TEMR, transposable element-mediated rearrangement; LINE; long interspersed nuclear element. b An example of TEMR-HR (top). Breakpoint junction of 1,294 bp TEMR deletion in NA19240 (middle) and alignment between flanking Alu elements to a Alu consensus sequence (bottom). c An example of TEMR-NHE (top). Breakpoint junction of 312 bp TEMR deletion in HG00514 (middle) and alignment between flanking Alu elements to a Alu consensus sequence (bottom). REF, reference genome (GRCh38). d The breakpoint microhomology distribution differs between NHE (orange) and HR (blue) TEMRs. e top: Microhomology GC content distribution for Alu TEMRs (dark green, n = 330), reference Alu elements (light green, n = 1,181,072), LINE-1 TEMRs (dark purple, n = 38), reference LINE-1s (light purple, n = 962,085) and the full human genome reference (HGR, gray). Bottom: Average GC content of TEMR breakpoint microhomologies for Alu (green) and LINE-1 (purple) TEMRs. Microhomologies were restricted to 5+ bp for this analysis. A two-sided Welch’s t-test was used to calculate the p-value. n.s, not significant.