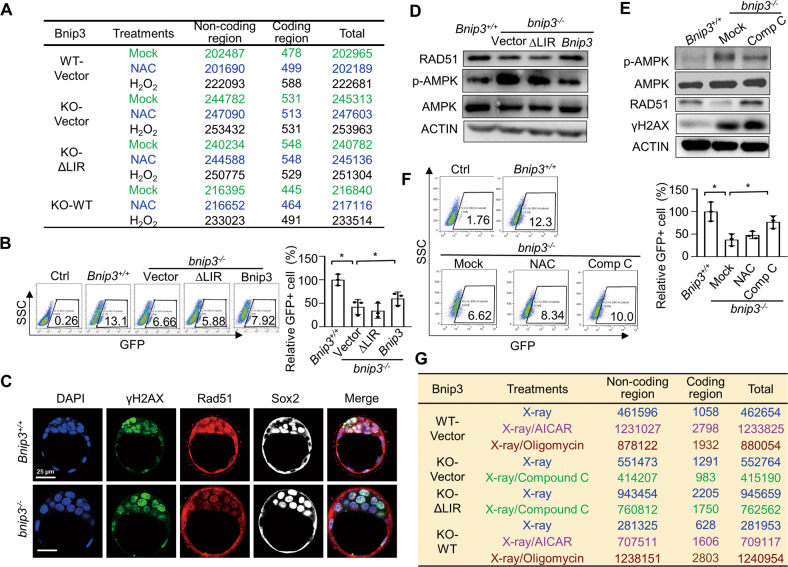
Fig. 3. BNIP3-dependent mitophagy safeguards homologous recombination.
A Acquired indels (insertions-deletions) in Vector-Bnip3+/+, Vector-binp3−/−, ∆LIR-binp3−/− and WT-binp3−/− ESC lines after 30 passages with the indicated treatments. Whole genome sequencing of Vector-Bnip3+/+, Vector-binp3−/−, ∆LIR-binp3−/−, and WT-binp3−/− ESC lines were performed and acquired indels were showed in table. Data shown are one representative of biological replicates. B Deteriorated HR in binp3−/− ESCs was partially rescued by wild-type but not mutant ∆LIR Bnip3. Data shown are the mean ± SD, n = 3; *, P < 0.05; Student’s t-test. C Decreased colocalization of γH2AX and RAD51 in the inner cell mass of bnip3−/− blastocysts. Green, γH2AX; Red, RAD51; White, SOX2; Blue, DAPI. D Excessive AMPK activation and decreased RAD51 expression in binp3−/− ESCs was partially rescued by wild-type but not ∆LIR Bnip3. E Inactivation of AMPK by Compound C restored compromised expression of RAD51 in binp3−/− ESCs without affecting γH2AX expression. Actin served as the loading control. F Compromised HR in binp3−/− ESCs was partially rescued by adding AMPK inhibitor Compound C, but not antioxidant NAC. Data shown are the mean ± SD, n = 3; *, P < 0.05; Student’s t-test. G Acquired indels in Vector-Bnip3+/+, Vector-binp3−/−, ∆LIR-binp3−/− and WT-binp3−/− ESCs with indicated treatments. Whole genome sequencing of Vector-Bnip3+/+, Vector-binp3−/−, ∆LIR-binp3−/−, and WT-binp3−/− ESC lines were performed and acquired indels were shown in table. Data shown are one representative of biological replicates.