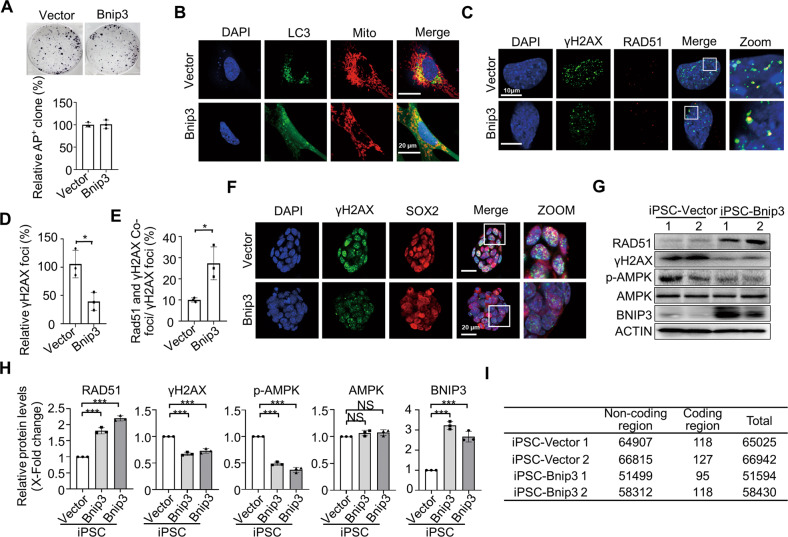
Fig. 4. Bnip3 overexpression improves iPSC genomic integrity.
A Overexpression of Bnip3 does not affect reprogramming efficiency. Data shown are the mean ± SD, n = 3; NS no significant difference. B Bnip3 overexpression enhanced mitochondrial autophagy in reprogramming cells. GFP: LC3; RFP: Mitochondria. C Confocal fluorescence micrographs of reprogramming cells. SSEA-1-positive cells at reprogramming day 3 were sorted for staining with γH2AX and RAD51. Green, γH2AX; Red, RAD51; Blue, DAPI. D Statistical data for γH2AX foci in (C). Cells were randomly divided into 3 groups and the foci were counted. Data shown are the mean ± SD, n = 3; *, P < 0.05; Student’s t-test. E Statistical data for RAD51 and γH2AX colocalization foci/γH2AX foci in (C). F Confocal fluorescence micrographs of iPSCs. Green, γH2AX; Red, SOX2; Blue, DAPI. G Western blot detection of RAD51, γH2AX, p-AMPK, AMPK, and BNIP3 in wild-type or Bnip3-overexpressing iPSCs. Actin served as the loading control. H Quantifications of each protein expression in (G). Data are shown as mean ± SD, n = 3; ***, P < 0.001; NS no significant difference, Student’s t-test. I Acquired indels in indicated iPSC lines.