Abstract
The adaptive potential of plant fungal pathogens is largely governed by the gene content of a species, consisting of core and accessory genes across the pathogen isolate repertoire. To approximate the complete gene repertoire of a globally significant crop fungal pathogen, a pan genomic analysis was undertaken for Pyrenophora tritici-repentis (Ptr), the causal agent of tan (or yellow) spot disease in wheat. In this study, 15 new Ptr genomes were sequenced, assembled and annotated, including isolates from three races not previously sequenced. Together with 11 previously published Ptr genomes, a pangenome for 26 Ptr isolates from Australia, Europe, North Africa and America, representing nearly all known races, revealed a conserved core-gene content of 57 % and presents a new Ptr resource for searching natural homologues (orthologues not acquired by horizontal transfer from another species) using remote protein structural homology. Here, we identify for the first time a non-synonymous mutation in the Ptr necrotrophic effector gene ToxB, multiple copies of the inactive toxb within an isolate, a distant natural Pyrenophora homologue of a known Parastagonopora nodorum necrotrophic effector (SnTox3), and clear genomic break points for the ToxA effector horizontal transfer region. This comprehensive genomic analysis of Ptr races includes nine isolates sequenced via long read technologies. Accordingly, these resources provide a more complete representation of the species, and serve as a resource to monitor variations potentially involved in pathogenicity.
Keywords: necrotrophic fungi, plant pathogen, toxin
Data Summary
The sources and genomic sequences used throughout this study have been deposited in the National Center for Biotechnology Information (NCBI), under the assembly accession numbers provided in Tables 1 and 2. The new resource for the Ptr isolate M4 protein structural homology is freely available through the BackPhyre web-portal: http://www.sbg.bio.ic.ac.uk/phyre2/.
Impact Statement.
Our Pyrenophora tritici-repentis (Ptr) pangenome study provides resources and analyses for the identification of pathogen virulence factors, of high importance to microbial research. Key approaches/findings include: (1) Analysis of 11 new sequenced (with three new races not previously available) and previously published isolates, 26 genomes in total, representing the near complete Ptr race set for known necrotrophic effector production collected from Australia, Europe, North Africa and the Americas. (2) We show that although Ptr has low core gene conservation, the whole genome divergence of other wheat pathogens was greater. (3) The new PacBio sequenced genomes provide unambiguous genomic break points for the large ToxA effector horizontal transfer region, which is only present in ToxA-producing races. (4) A new web-based resource for searching Ptr remote protein structural homology in silico is presented, and for the first time distant natural Pyrenophora protein homologues are identified to the Parastagonopora nodorum necrotrophic effector SnTox3.
Introduction
Tan (or yellow) spot, caused by the necrotrophic fungal pathogen Pyrenophora tritici-repentis [(Died.) Drechs.] (Ptr), can occur on both bread wheat (Triticum aestivum L.) and durum wheat (T. turgidum subsp. durum L.). A globally significant disease of economic importance [1, 2], tan spot can reduce crop production, with up to 31 % yield losses reported [3].
During infection, necrotrophic fungi secrete necrotrophic effectors (NEs) that interact with the corresponding sensitivity genes in the host wheat lines [4–7]. To date, Ptr has three known NEs, Ptr ToxA, Ptr ToxB and Ptr ToxC, that lead to either necrosis or chlorosis symptoms on their sensitive wheat genotypes [8, 9]. It is the different combinations (and absence) of these NEs that have been used to define different Ptr races [6, 10], including race 1 (Ptr ToxA and Ptr ToxC), race 2 (Ptr ToxA), race 3 (Ptr ToxC), race 4 (no Ptr ToxA, Ptr ToxB or Ptr ToxC), race 5 (Ptr ToxB), race 6 (Ptr ToxB and Ptr ToxC), race 7 (Ptr ToxA and Ptr ToxB) and race 8 (Ptr ToxA, Ptr ToxB and Ptr ToxC). However, there are reports of isolates beyond the current classification [1, 11, 12]. AR CrossB10 from North Dakota, USA, was such an isolate that produces both Ptr ToxC with an unknown NE, which has been recently sequenced [13] and will subsequently be referred to here as ‘race unknown’ [14]. In addition to the three known NEs, the presence of novel NEs has been suggested in several studies [11, 15–18]. Pathogenic fungi possess large effector repertoires that comprise hundreds of small secreted proteins only related by protein tertiary structures (3D structure) [19]. The detection of novel NEs makes the sequencing of new isolates a priority to capture and understand the complete gene repertoire for Ptr and highlights the importance of in silico screening of known protein tertiary structures to identify new NE protein families.
Genome sequencing projects for fungal pathogens using single molecule long reads, such as PacBio and Oxford Nanopore technologies, have significantly improved our understanding of pathogen genomes, as they allow near complete genome assembly. In particular, the wheat fungal pathogens Fusarium graminearum (cause of fusarium head blight), Ptr, Parastagonospora nodrum (Sn, cause of Septoria nodorum blotch) and Zymoseptoria tritici (Zt, cause of Septoria tritici blotch) are known to have highly variable genomes characterized by gene loss and duplication events as well as large-scale genome rearrangements [14, 20–24]. To understand the genome composition of a species, the protein-coding genes from all available isolates are clustered based on the sequence identity of conserved protein domains into core (genes shared by all isolates) and accessory (genes absent in one or more isolates) groups. The union of the core and accessory groups for the collection of isolates is then referred to as the pangenome, which is larger than the genome of any one individual [25]. Depending on the number of sequenced isolates, associations with distinct habitats and phenotypes may then be detected within a pathogen species [25].
In this study, 15 new Ptr isolates collected from Europe (Denmark, Germany and the UK), North Africa (Algeria and Tunisia) and the Americas (Brazil, Canada and the USA) were sequenced, assembled and annotated, for comparative analysis with 11 previously published Australian and North American Ptr isolates [14, 23, 26]. A total of 26 annotated Ptr genomes, which represent nearly all known Ptr races, are presented here for a pangenome analysis to determine whole genome phylogeny and sequence variations in relation to core and accessory genes. Ptr proteins are then further explored in silico to identify remote natural structural homology between different necrotrophic fungal species (not acquired by horizontal gene transfer).
Methods
Isolate collection and DNA extraction
Ptr isolates were collected from Algeria (Alg130 and Alg215), Brazil (Biotrigo9-1), Canada (90-2), Denmark (EW306-2-1, EW4-4 and EW7m1), Germany (SN001A, SN001C and SN002B), USA (86-124 and Ls13-192), UK (CC142) and Tunisia (T199 and T205). All isolates were collected from bread wheat (T. aestivum L.), except Alg215 which was collected from durum wheat (T. turgidum subsp. durum L.). Fungi were grown on V8-PDA agar as described [27]. Genomic DNA was extracted using a BioSprint 15 DNA Plant Kit (Qiagen) with some modifications. Briefly, DNA was extracted using the BioSprint 15 automated workstation, according to the manufacturer’s instructions, from 3 day old mycelia grown in Fries 3 medium [27]. DNA was further treated with 50 µg ml−1 of RNase enzyme (Qiagen) for 1 h followed by phenol/chloroform extraction, precipitation with sodium acetate and ethanol, and finally resuspension in Tris-EDTA buffer.
Isolate pathotyping
Ptr isolates were pathotyped for race classification through infection assays of differential wheat genotypes differing in their specific NE sensitivities. The wheat genotypes used were Glenlea (Ptr ToxA-sensitive), 6B662 (Ptr ToxB-sensitive), 6B365 (Ptr ToxC-sensitive), and Auburn or Salamouni (insensitive to all three NEs).
Two-week-old wheat (T. aestivum L.) seedlings were inoculated by spraying conidia onto the whole plants evenly at a rate of 3 000 conidia ml–1 and grown at 20 °C under a 12 h day/night cycle in a controlled growth chamber [27]. The second leaves were harvested 7 days post-inoculation, visually inspected for symptoms [28] and photographed. The inoculation experiments were repeated twice with three replicate plants per wheat genotype.
Ptr isolate sequencing and genome assembly
Genomic DNA from four Ptr isolates was sequenced using the PacBio Sequel system, 90-2 (Novogene), Biotrigo9-1 (Novogene), Ls13-192 and 86-124 (Mayo Clinic). Error correction and de novo genome assembly of PacBio reads was completed with Canu version v2.1.1 [29] with the following options (genomeSize=43, useGrid=TRUE, maxThreads=28, merylThreads=28, ovlThreads=28 ovlMerThreshold=500 and gridOptionsOBTOVL="--cpus-per-task=28) on computer resources (Broadwell Intel Xeon cores, 100 Gb s–1 Omni-Path interconnect and 128 GB of memory per compute node) at Pawsey Supercomputing Centre, Perth, Western Australia. Previously generated Illumina 150 bp paired-end DNA sequence reads of 86-124 genomic DNA [14] and 90-2 and Biotrigo9-1 Illumina sequence (this study) were aligned to the contigs using BWA V0.7.17-r1188 [30], and the sorted alignment bam files then used for further base error corrections (one round) using Pilon v1.24 [31]. The fraction of errors corrected were 1.4e-05, 1.6e-05 and 1.6e-06 for Biotrigo9-1, 86-124 and 90-2, respectively.
The genomic DNA for an additional 11 Ptr isolates (EW306-2-1, EW4-4, EW7m1, SN001A, SN001C, SN002B, CC142, Alg130, Alg215, T199 and T205) was sequenced (at 100× genome coverage) using Illumina Hi-Seq 150 bp pair-end reads by the Australian Genome Research Facility (AGRF). Isolate sequence data was quality checked with FASTQC [32], and trimmed for poor quality, ambiguous bases and adapters using Skewer [33] and Trimmomatic v0.22 [34] with a read head crop of 6 bp and minimum length of 100 bp. De novo genome assembly was undertaken using SPAdes version v3.10.0 with --cov-cutoff auto and --careful options [35].
Gene prediction and functional annotation
Ptr sequenced genomes were soft masked for low complexity, as well as known transposable elements, using RepeatMasker (RM) [36] v. open-4.0.6 with rmblastn version 2.2.27+ on RepBase [37] RM database version 20 150 807 (taxon=fungi). Ab initio gene predictions were made with GeneMark-ES v4.33 (--ES --fungus --cores 16) [38] and CodingQuarry v1.2 Pathogen Mode (PM) [39], assisted by RNA-Seq [40] genome alignments using TopHat2 [41] for a minimum intron size of 10 bp. The Ptr M4 and Pt-1C-BFP reference proteins [14, 23] were aligned using Exonerate v2.2.0 (--minintron 10 --maxintron 3000) protein2genome mode [42]. Gene annotations were assigned from blastx (v2.2.26) [43] searches against the Uniref90 (October 13 2020), NCBI Refseq (taxon=Pezizomycotina) (October 13 2020) and InterProScan v5.17-56 [44] protein databases. Sequence domains were assigned by RPS-blast (v2.2.26) against the Pfam v33.1, Smart v6.0 and CDD v3.19 databases. The blast protein and domain searches were then summarized using AutoFACT v3.4 [45].
Proteins were screened for a signal peptide using SignalP v5.0b [46]. Effector predictions were made on proteins with signal peptides using EffectorP v3.0 [47, 48]. To ensure the same prediction methods were used for comparative analyses, SignalP V5.0b and EffectorP v3.0 [47, 48] were used to update the effector gene predictions on all the publicly available isolate genomes (Supplementary data 1). All predicted proteins were also ranked using Predector v1.1 [49] (Supplementary data 1). Gene completeness was accessed using BUSCO v3, lineage fungi [50].
Comparative genomics
To conduct comparative analyses across the class Ascomycota, publicly available isolate genomes were downloaded from the National Center for Biotechnology Information (NCBI) GenBank. These included Bipolaris (B. cookei, B. maydis, B. sorokiniana, B. zeicola), Leptosphaeria (L. maculans), Parastagonospora (P. nodorum), Pyrenophora (Pyrenophora teres f. teres, Pyrenophora teres f. maculata, Pyrenophora serminiperda) and Zymoseptoria (Z. tritici), [21, 24, 51–53] (Supplementary data 2). The published genomes of P. tritici-repentis isolates Pt-1C-BFP, DW5, DW7, SD20 [23], Ptr134, Ptr239, Ptr11137, Ptr5213, M4, 86-124 [14], AR CrossB10 [13] and V1 [26] were also included for analysis.
Genome nucleotide pairwise distance was calculated with Phylonium v1.5 [54] with two-pass enabled and 100 bootstrap matrices. Whole genome phylogenetic trees were reconstructed using Phylip 1 : 3.695-1 [55], consensus program v3.695 on 100 Kitsch and neighbour-joining v3.695 trees. The tree was then visualized using FigTree v1.4.4. Genomic nucleotide regions were compared between isolates using NUCmer v3.1 [56] and Easyfig v2.2.3 [57].
To determine the presence, copy number and percentage identity of all genes in Ptr, the gene nucleotide sequences from all 26 isolates were aligned to all 26 genomes using GMAP version 2021-05-27 with options ‘-f 2 t 48 n 300 --max-intronlength-middle=3000 --max-intronlength-ends=3000 --fulllength --trim-end-exons=0 --alt-start-codons --canonical-mode=1 --max-deletionlength=20’. Isolate coding sequence (CDS) Pearson correlations and predicted effector protein lengths and scores were analysed using R v4.0.3 [58] using the R packages corrplot v0.84, ggplot2 v3.3.3, ggridges v0.5.3 and pheatmap v1.0.12. The analysis and data are available in v1.3.1093 RStudio [59] markdown notebook: https://github.com/ccdmb/PTR-60.
Isolate reads were aligned to the isolate M4 reference genome using BWA 0.7.14-r1138, and coverage (10 kb windows) was calculated using BedTools (genomecov) v2.17.0 on SamTools v0.1.19-96b5f2294a sorted bam files. Regions of absence were then plotted using Circos v0.69-3 and R v3.5.1, bioconductor package chromPlot v1.10.0.
Protein orthologous clustering and effector analysis
Predicted protein data for all available Ptr isolates were clustered using OrthoFinder v2.5.2 [60]. Orthogroup protein sequences for were aligned using muscle v3.8.1551 [61] and a consensus sequence created using consambig EMBOSS v6.6.0.0 [62]. The consensus sequences and singletons were then searched against v35.0 of Pfam Hidden Markov Models (HMMs) [63] using hmmsearch v3.3 [64] with an expected value threshold of 1e-05. Gene ontologies were then assigned using pfam2go v2022/03/16 [65]. The predicted effector groups (with signal peptides) were then screened for three-dimensional (3D) protein model predictions using the Protein Homology/analogY Recognition Engine V2.0 Phyre2 [66] batch processing mode. The predicted models were superimposed on the best ranked template to find the largest subset of atoms within an approximate threshold of 3.5 Å, which was adjusted based on the size of the aligned proteins using iMol [67]. Protein sequences with high confidence (Phyre2 ≥90 %) predicted 3D protein models were also searched against the Plant Host Interactions database (PHI-base) of known pathogenic phenotypes [68], at an expected value threshold of ≤1e-10 for significant alignments. HMM libraries were created for the whole genome of Ptr isolate M4, which has been made publicly available through the online resource BackPhyre, Imperial College, London [66].
Results
PacBio genome sequencing, assembly and annotation of four Ptr isolates
A total of four Ptr genomes comprising two race 4 (lacking all three known Ptr NEs) isolates [North Dakota (USA) isolate Ls13-192 [69] and Canadian isolate 90-2 [70], and two race 2 (producing Ptr ToxA only) isolates (Brazilian isolate Biotrigo9-1 [71] and Canadian isolate 86124 [72]) were sequenced using PacBio technology, assembled and protein-coding genes were predicted for comparative analysis.
The assembled Ptr genomes ranged in size from 37.56 to 42.19 Mb (Table 1) and, of these, the known NE-producing isolates (86-124 and Biotrigo9-1) had a size comparable to previously PacBio-sequenced genomes (M4 and DW5) [14, 73]. The race 4 isolate not producing known NEs, Ls13-192, had the smallest genome size at 37.56 Mb, at least 2 Mb smaller than all the known NE-producing isolate genomes, but similar to Pt-1C-BFP, which was sequenced prior to the availability of third-generation long read technologies and which lacks some representation of repeat/complex genomic regions [23]. Our four new assemblies were more fragmented than the previously assembled genomes M4 and DW5 [14, 73]. In particular race 4 isolate 90-2 was fragmented into 162 contigs, over twice as many contigs as compared to race 2 isolate Biotrigo9-1 and race 4 isolate Ls13-192. The four genome assemblies had a BUSCO quantitative assessment >98.9 % for completeness with respect to gene content (Fig. S1).
Table 1.
Summary statistics for our four PacBio sequenced Ptr genome assemblies, compared with those of two previously published Ptr assemblies
|
86-124 |
Biotrigo9-1 |
Ls13-192 |
90-2 |
M4* |
DW5* |
|
|---|---|---|---|---|---|---|
|
Race |
2 |
2 |
4 |
4 |
1 |
5 |
|
Known NE |
A |
A |
– |
– |
AC |
B |
|
toxb |
toxb x2 |
|||||
|
Source |
Canada |
Brazil |
USA |
Canada |
Australia |
USA |
|
GenBank accession |
NRDI02 |
JAHCYZ00 |
JAHCSW00 |
JAAFOX00 |
NQIK02 |
MUXC02 |
|
Genome coverage |
200× |
200× |
200× |
164× |
100× |
77× |
|
Number of contigs |
139 |
75 |
72 |
162 |
50 |
60 |
|
Total contig length (Mb) |
41.15 |
42.19 |
37.56 |
39.71 |
40.92 |
40.87 |
|
Mean contig size (kb) |
296.04 |
562.57 |
521.68 |
245.18 |
998.09 |
681.19 |
|
Median contig size (bp) |
23 098 |
34 534 |
32 389 |
20 792 |
32 745 |
31 213 |
|
Longest contig (Mb) |
3.92 |
10.08 |
7.54 |
7.30 |
9.91 |
8.11 |
|
Shortest contig (bp) |
3 180 |
8 676 |
1 765 |
2 050 |
3 304 |
2 843 |
|
Contigs >10 kb† |
113 (81.29 %) |
74 (98.67 %) |
69 (95.83 %) |
152 (93.3 %) |
38 (92.68 %) |
39 (65.00 %) |
|
Contigs >100 kb† |
41 (29.50 %) |
18 (24.00 %) |
18 (25.00 %) |
39 (24.07 %) |
11 (26.83 %) |
17 (28.33 %) |
|
Contigs >1 Mb† |
14 (10.07 %) |
12 (16.00 %) |
12 (16.67 %) |
9 (5.56 %) |
10 (24.39 %) |
12 (20.00 %) |
|
N50 |
1 684 023 |
3 177 932 |
2 530 800 |
1 794 835 |
3 658 030 |
3 133 851 |
|
L50 |
9 |
5 |
5 |
5 |
4 |
5 |
|
N80 |
623 938 |
1 969 426 |
1 691 594 |
567 608 |
2 765 034 |
2 129 786 |
|
L80 |
21 |
10 |
10 |
17 |
8 |
10 |
|
Genes |
14 272 |
14 450 |
12 851 |
14 224 |
15 459 |
14 276 |
|
Total protein (aa) length (Mb) |
6.94 |
7.01 |
6.04 |
5.86 |
6.90 |
5.95 |
|
Predicted effectors‡ |
178 (1.2 %) |
169 (1.1 %) |
189 (1.4 %) |
380 (2.6 %) |
291 (1.8 %) |
314 (2.1 %) |
*Previously published in NCBI GenBank. Necrotrophic effector (NE).
†Percentage of contigs over the displayed length is shown in parentheses.
‡EffectorP V3 predictions ≥0.7, the percentage of genes predicted in an effector is shown in parentheses. N50 and N80 is the sequence length of the shortest contig at 50 and 80 % of the total genome length. L50 and L80 is the count of the smallest number of contigs whose length sum makes up 50 and 80 % of the genome size, respectively.
The number of predicted protein-encoding genes for our new PacBio-sequenced genome assemblies ranged from 12 851 (Ls13-192) to 14 450 (Biotrigo9-1) (Table 1). The number of predicted protein effectors for race 2 isolates 86-124 and Biotrigo9-1 and race 4 isolate Ls13-192 was lower than the numbers predicted for race 1 solate M4 and race 5 isolate DW5. However, race 4 isolate 90-2 had the highest number of proteins predicted as effectors, due to a higher gene copy number identified later in the protein clustering analysis. Furthermore, in the race 4 isolates a single toxb (found in non-pathogenic Ptr isolates and having no toxic activity [74, 75]) was detected in Ls13-192 on contig 4 (113 627–113 893 bp) and an exact toxb duplication event was detected in 90-2 on contig 37 (termed here toxb1, 15 199–15 465 bp) and on contig 42 (termed toxb2, 15 135–15 401 bp). The toxb genes appeared close to a contig end. Ls13-192 contig 4 and 90-2 contigs 37 and 42 have contig assembly sizes 3 110 kb, 116 kb and 87 kb, respectively. No toxb gene coding region, protein or nucleotide sequence variations were identified (Figs S2 and S3). ToxA was identified in race 2 isolates 86-124 (contig 17, 764 135–764 722 bp) and Biotrigo9-1 (contig 7, 1 370 173–1 370 760 bp), but no gene coding region, nucleotide or protein sequence variations were found. The Ptr-specific hairpin element (PtrHp1) ToxA 3′ UTR insertion previously identified) [76] was not detected in the ToxA 3′ UTR region of these genomes.
The four new assembled and annotated genomes Ls13-192, 86-124, 90-2 and Biotrigo9-1 have been deposited in NCBI GenBank and can be found under accession numbers JAHCSW000000000, NRDI02000000, JAAFOX000000000 and JAHCYZ000000000, respectively.
Illumina genome sequencing, assembly and annotation of 11 Ptr isolates
Whole genome Illumina sequencing and assembly was then undertaken for 11 new Ptr genomes consisting of isolates from Denmark (EW306-2-1, EW4-4 and EW7m1), Germany (SN001A, SN001C and SN002B), UK (CC142), Algeria (Alg130 and Alg215) and Tunisia (T199 and T205). The assembled Ptr genomes ranged in size from 34.15 to 35.18 Mb (Table 2), comparable to previous Illumina Ptr isolate assembly sizes [14].
Table 2.
Illumina sequenced genome assemblies of 11 new Ptr isolates: isolate source, race and de novo assembly statistics
|
Isolate |
CC142 |
EW306-2-1 |
EW4-4 |
EW7m1 |
SN001A |
SN001C |
SN002B |
Alg130 |
T199 |
T205 |
Alg215 |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
GenBank accession |
|||||||||||
|
Source |
UK |
Denmark |
Denmark |
Denmark |
Germany |
Germany |
Germany |
Algeria |
Tunisia |
Tunisia |
Algeria |
|
Year collected |
– |
2015 |
2015 |
2015 |
2016 |
2016 |
2016 |
2016 |
2016 |
2016 |
2015 |
|
Race |
1 |
1 |
1 |
3 |
3 |
nd |
nd |
5 |
7 |
4 |
8* |
|
Known NEs |
ToxA, ToxC |
ToxA, ToxC |
ToxA. ToxC |
ToxC |
ToxC |
ToxA† |
ToxA† |
ToxB |
ToxA, ToxB |
None |
ToxA, ToxB‡, ToxC |
|
Locus ID |
10 965 |
03 130 |
05 320 |
– |
– |
12 604 |
05700 |
11 547 |
11 003; 11 565 |
05 415 |
|
|
ToxA-PtrHp1 |
Present |
Absent |
Present |
Absent |
Absent |
Present |
Absent |
Absent |
Absent |
Absent |
Absent |
|
Contigs |
2 398 |
2 590 |
2 353 |
2 406 |
2 367 |
2 483 |
2 952 |
3 407 |
3 173 |
3 079 |
3 134 |
|
Total length (Mb) |
34.34 |
34.54 |
34.36 |
34.22 |
34.15 |
34.29 |
35.15 |
34.97 |
34.43 |
34.28 |
34.72 |
|
Mean contig size |
14 322 |
13 336 |
14 606 |
14 226 |
14 428 |
13 811 |
11 910 |
10 266 |
10 852 |
11 136 |
11 335 |
|
Longest contig |
205 419 |
291 678 |
233 712 |
233 813 |
258 315 |
188 750 |
233 762 |
272 310 |
309 714 |
272 252 |
342 080 |
|
N50 |
47 343 |
48 368 |
48 593 |
48 975 |
49 129 |
45 477 |
48 749 |
53 052 |
55 190 |
54 028 |
63 004 |
|
L50 |
213 |
202 |
206 |
199 |
202 |
221 |
216 |
187 |
174 |
179 |
159 |
|
Genes |
12 348 |
12 498 |
12 323 |
12 427 |
12 311 |
12 388 |
12 472 |
12 387 |
12 256 |
12 172 |
12 475 |
|
Total CDS length (Mb) |
15.72 |
15.79 |
15.72 |
15.66 |
15.62 |
15.70 |
15.76 |
16.24 |
16.14 |
16.09 |
16.34 |
|
Predicted effectors§ |
279 (2.25 %) |
289 (2.31 %) |
287 (2.32 %) |
284 (2.28 %) |
281 (2.28 %) |
287 (2.31 %) |
291 (2.33 %) |
300 (2.42 %) |
297 (2.42 %) |
286 (2.34 %) |
291 (2.33 %) |
*Provisionally assigned as race 8. †Not determined; colonies were not viable for spore production. ‡Partial sequence that is truncated and contains a synonymous SNP. §EffectorP3.0 score ≥0.7, the percentage of genes predicted in an effector is shown in parentheses. Necrotrophic effectors (NEs).
The number of predicted protein-encoding genes ranged between 12 172 and 12 498 for the assembled genomes. Of these, 279–300 effectors were predicted with a probability score ≥0.7. ToxA was identified in isolates T199, Alg215, CC142, EW3061-2-1, EW4-4, SN001C and SN001B, and ToxB was identified in the Alg130 genome. ToxA and ToxB were both detected in T199 and Alg215 genomes, but the Alg215 ToxB sequence was partial (due to a sequence inversion in the 5′ end of the gene), truncated by 33 amino acid residues in the protein N terminus which includes the encoded signal peptide (amino acid positions 1–22) (Fig. S4). Furthermore, a single non-synonymous substitution (I→R, residue position 17) was detected. Neither ToxA nor ToxB were detected in isolates EW7m1, SN001A and T205.
The Ptr-specific hairpin element (PtrHp1) ToxA 3′ UTR insertion previously identified in isolates EW306-2-1 and EW4-4 [76] was also detected in ToxA 3′ UTR for our UK isolate CC142, but not in the remaining North African ToxA isolates T199 and Alg215 (Table 2).
The plant infection assays on the wheat differential lines confirmed CC142, EW306-2-1 and EW4-4 as race 1 isolates (producing ToxA and ToxC), EW7m1 and SN001A as race 3 isolates (producing ToxC), Alg130 as a race 5 isolate (producing ToxB), T199 as a race 7 isolate (producing ToxA and ToxB) and T205 as a race 4 isolate (no ToxA, ToxB or ToxC production) (Figs S5 and S6). Due to the truncated ToxB gene in isolate Alg215 and a weaker chlorosis phenotype on the ToxB wheat differential lines, Alg215 has been provisionally classified as a race 8 isolate (producing ToxA, ToxB and ToxC) (Table 2). The SN001C and SN002B isolates could not be tested for race classification because the colonies sporulated poorly; nonetheless, ToxA was present and ToxB was absent in the genome sequence for both isolates. As ToxC production in SN001C and SN002B remains unknown, they could be race 1 or 2.
All the assembled and annotated genomes have been deposited in NCBI GenBank and can be found under accession numbers PSOO00000000–PSOU00000000 and RXHK00000000–RXHN00000000.
Whole genome comparative analyses
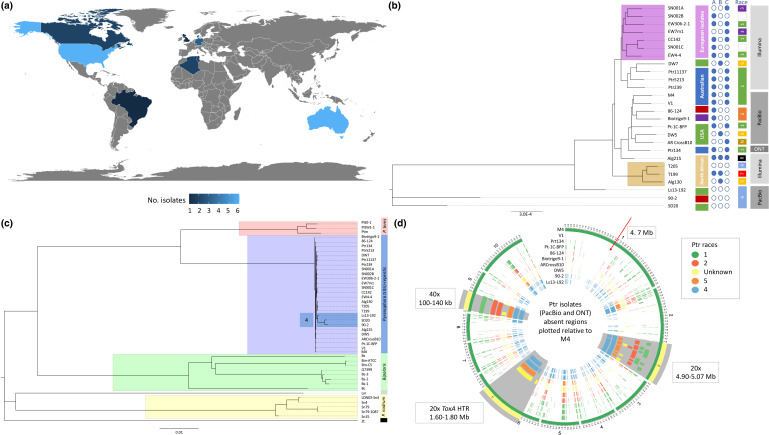
Whole genome phylogenetic analysis of the 26 Ptr isolates, sourced from the major wheat-growing regions in the Americas, Australia, Europe and North Africa (Fig. 1a), showed distinct clades for European and North African geographical locations (Fig. 1b). Surprisingly, isolate Alg215 from North Africa did not cluster with the remaining North African isolates. On genome alignment to the reference genome of isolate M4, a large 1 Mb distal region on M4 contig 1 and many smaller regions were absent in Alg130, T199 and T205 but were present in Alg215 (Fig. S7). Furthermore, branches for race 4 (that do not produce ToxA, ToxB or ToxC) isolates (SD20, 90-2 and Ls13-192) had the greatest phylogenetic distances from the known NE-producing isolate groups, while race 4 T205 and SD20 (both Illumina sequenced) did not cluster. In particular, isolates SD20 (USA) and 90-2 (Canada) were more distant than the isolate Ls13-192 (USA).
Fig. 1.
Whole genome analysis of Ptr isolates. (a) Geographical source and number of Ptr isolates currently available and analysed. Key gives the number of isolates. (b) Whole genome phylogenetic tree of Ptr isolates from Illumina sequencing (Alg130, T199, T205, Alg215, CC142, EW306-2-1, EW4-4, EW7m1, DW7, Pt-1C-BFP, Ptr239, Ptr11137, Ptr5213, SD20, SN001A, SN001C, SN002B), PacBio Technologies (86-124, 90-2, AR CrossB10, Biotrigo9-1, DW5, Ls13-192, M4 and V1) and Oxford Nanopore Technologies (Ptr134). The unrooted neighbour-joining phylogenetic tree displays clades for the European (violet) and North Africa (tan) isolates. Geographical source of the other isolates is Australia (blue), USA (green), Canada (red) and Brazil (purple). The race 4 isolates (Ls13-192, 90-2 and SD20) have the greatest distance from the clade of known NE-producing isolates. (c) Unrooted neighbour-joining phylogenetic tree for Ptr (purple clade), Pyrenophora teres [P. teres f. maculata (Ptm) and P. teres f. teres (Ptt)) (orange clade), Bipolaris [B. sorokiniana (Bs1-3 and Q7399), B. maydis (Bm-ATCC and Bm-C5) and Bipolaris zeicola (Bz)] (green clade), Parastagonospora nodorum (Sn4, Sn15 and Sn79) (yellow clade), Leptosphaeria maculans (Lm) and Zymoseptoria tritici (Zt) isolates. The branches for race 4 isolates not producing known NEs (Ls13-192, 90-2 and SD20) are highlighted (blue) within the Ptr clade. (d) Circular plots show 10 kb regions of absence plotted for the Ptr isolate genomes sequenced using long-read technologies (PacBio and Oxford Nanopore Technology) as compared with the chromosomes of the reference Ptr genome of isolate M4. Isolates are coloured by race. Three regions of interest are highlighted in grey and zoomed at 20× for chromosome 2 and chromosome 1, and 40× for chromosome 9.
Whole genome phylogenetic analysis of Ptr and related ascomycete fungal species clustered into four distinct clades for Bipolaris species, P. nodorum, P. tritici-repentis and P. teres (Fig. 1c). A lower phylogenetic divergence within the individual Pyrenophora species (P. teres f. maculata (Ptm), P. teres f teres (Ptt) and Ptr) was observed as compared with Bs, Pn and Zt isolates (Fig. S8).
To observe regions of absence across the assembled genomes, regions ≥10 kb absent for the Ptr isolates were plotted against the reference M4 genome (Fig. 1d). The large horizonal transferred region for ToxA on chromosome 6 (chr6) was present in all ToxA-producing isolates and absent in ToxA non-producing isolates. For the previously reported large Ptr ToxA horizontal transfer region, believed to have come from P. nodorum [14, 23, 77], clear break points on M4 chr6 at positions 1 645 874 and 1 774 022 bp (128 kb insertion) could be determined between isolates producing and not producing ToxA (Figs 1d and S9). The flanking regions of the breakpoints were highly conserved between all the aligned isolates (Fig. S9). A region on chr1 near the 1.47 Mb position was absent in all non-ToxC-producing isolates and the unknown race (ToxC-producing) when only looking at long read assemblies [Figs 1d and S10 (plot on left hand side)]. The race 4 isolates had more regions of absence, particularly in the distal ends of chr2. A greater number of absent regions was obtained for Illumina-sequenced assemblies (Fig. S10, plot on right hand side). Regions of variation appear mostly associated with chromosome telomeres and centromeres. In particular, the distal region on M4 chr10, the equivalent of race 5 isolate DW5 chr11 (1 752 563–2 152 826 bp), was mostly unique as compared with races 1, 2, 4 and the unknown race, with fragmented alignments dispersed throughout the last 100 kb of the chromosome surrounding Ptr ToxB2 (2 152 563–2 152 826 bp) (Fig. S11).
Ptr cDNA sequence alignment to whole genomes
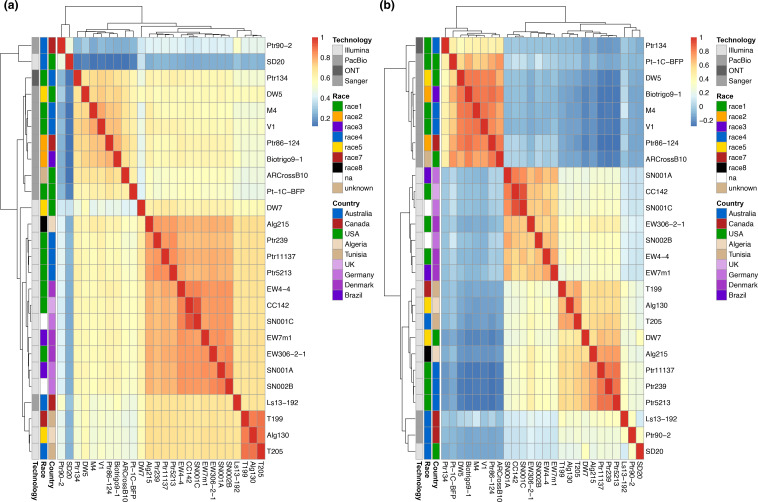
To ensure a comprehensive search of Ptr genes in the pangenome, predicted coding sequence (cDNA) sequences from all isolates were aligned to all the genomes at >90 % sequence identity and 90 % coverage. The number of alignments and greatest percentage identity for each locus were recorded to determine isolate correlations (Fig. 2). Although a closer correlation by gene percentage sequence identity could be determined for isolates that were Illumina- or PacBio-sequenced, a distinct grouping for Alg130, T199 and T205, and a grouping of the European isolates, was evident. Furthermore, the race 4 isolates 90-2 and SD20 were less correlated to all the remaining isolates (Fig. 2a). Based on gene counts (copy number), three distinct groups were observed, for long read-sequenced, European Illumina-sequenced and Australian/North African/North American Illumina-sequenced isolates (Fig. 2b). However, the three race 4 isolates (Ls13-192, 90-2 and SD20) were outliers.
Fig. 2.
Ptr pangenome predicted cDNA correlation plots for gene sequence percentage identity (a) and gene copy number (b). Ptr isolates from Illumina sequencing (Alg130, T199, T205, Alg215, CC142, EW306-2-1, EW4-4, EW7m1, DW7, Pt-1C-BFP, Ptr239, Ptr11137, Ptr5213, SD20, SN001A, SN001C, SN002B), PacBio Technologies [86-124 (Ptr86-124), 90-2 (Ptr90-2), AR CrossB10, Biotrigo9-1, DW5, Ls13-192, M4 and V1] and Oxford Nanopore Technologies (Ptr134).
All genes were then filtered for presence/absence variation between ToxC-producing isolates [race 1 (Pt-1C-BFP, CC142, EW4_4 and EW306-2-1), race 3 (EW7m1 and SN001A), race unknown (AR CrossB10) and provisional race 8 (Alg215)] and non-ToxC-producing isolates [race 2 (86-124 and Biorigo9-1), race 4 (T205, Ls13-192, 90-2 and SD20), race 5 (DW5 and DW7) and race 7 (T199)] to identify genes that may be related to ToxC production. When only PacBio-sequenced genomes were queried, a cluster of 16 genes from isolate M4 cDNAs 12 743–12 761 (proteins KAF7566087–KAF7566105) positioned on M4 chr9 within 101 367–138 426 bp and 15 single locus genes outside of the cluster were found present in the ToxC-producing races (races 1 and unknown) and absent in races not producing ToxC (races 2, 4 and 5) (Fig. S12). However, the region was absent for the race 1 Oxford Nanopore technology (ONT)-sequenced isolate Ptr134 (Fig. 1d). None of the 31 genes found to be specific to ToxC-producing isolates (based on PacBio technology) had an identified signal peptide or appeared to be part of any predicted biosynthetic gene cluster (Table 3 and Supplementary data 3). A search of the pathogen–host interaction database PHI-base [68], which provides expertly curated molecular and biological information on genes proven to affect the outcome of pathogen–host interactions, however, did identify four proteins with significant alignments to proteins with classified reduced virulence and lethal phenotypes. The following proteins with reduced virulence phenotype were described as being a Tfo1 transposon in Beauveria bassiana ARSEF 2860 (J4UFF8), a non-ribosomal protein synthase (NRPS) (A0A024CHY2) in Pseudomonas cichorii and an AMP binding protein (E3QPY3) in Colletotrichum graminicola. The protein I1RXA5, classified with a lethal phenotype in F. graminearum, appears to be a transcription factor (homeobox).
Table 3.
Ptr predicted cDNA sequences identified specific to ToxC-producing isolates (PacBio-sequenced) and PHI-base results
|
M4 GenBank accession |
Chr. |
Strand |
Gene position |
M4 cDNA |
Protein ID |
Description |
Length (aa) |
PHI base ID |
Expected value |
|---|---|---|---|---|---|---|---|---|---|
|
Chr1 |
+ |
4192655–4193250 |
mRNA_1649 |
Hypothetical protein |
161 |
||||
|
Chr2 |
− |
472148–474190 |
mRNA_3999 |
Dimer-Tnp-hAT domain-containing protein |
681 |
J4UFF8* |
4e-50 |
||
|
Chr2 |
− |
5052985–5053659 |
mRNA_5712 |
Hypothetical protein transmembrane |
224 |
||||
|
Chr3 |
+ |
94019–97874 |
mRNA_5765 |
Dimer-Tnp-hAT domain containing protein |
1104 |
A0A024CHY2* |
6.00E-18 |
||
|
Chr3 |
+ |
104828–106828 |
mRNA_5768 |
Hypothetical protein |
666 |
E3QPY3* |
2.00E-56 |
||
|
Chr3 |
− |
1062456–1062785 |
mRNA_6121 |
Hypothetical protein |
109 |
||||
|
Chr3 |
+ |
1234950–1235279 |
mRNA_6173 |
Hypothetical protein |
109 |
||||
|
Chr5 |
+ |
1432573–1432928 |
mRNA_8853 |
Hypothetical protein |
99 |
||||
|
Chr5 |
+ |
1504991–1506668 |
mRNA_8883 |
Hypothetical protein |
518 |
||||
|
Chr5 |
+ |
3 350 956.3351804 |
mRNA_9590 |
DDE-3 multi-domain protein |
282 |
I1RXA5† |
1.00E-55 |
||
|
Chr6 |
− |
157070–158024 |
mRNA_9652 |
Hypothetical protein |
260 |
||||
|
Chr6 |
− |
227855–229037 |
mRNA_9675 |
Hypothetical protein |
292 |
||||
|
Chr6 |
+ |
1257276–1261804 |
mRNA_10029 |
Hypothetical protein |
1489 |
||||
|
Chr9 |
− |
101367–102082 |
mRNA_12743 |
Hypothetical protein |
166 |
||||
|
Chr9 |
+ |
103558–104538 |
mRNA_12744 |
Hypothetical protein |
326 |
||||
|
Chr9 |
− |
108927–109433 |
mRNA_12746 |
Hypothetical protein |
153 |
||||
|
Chr9 |
− |
109617–109871 |
mRNA_12747 |
Hypothetical protein |
84 |
||||
|
Chr9 |
− |
112305–113390 |
mRNA_12748 |
Hypothetical protein |
346 |
||||
|
Chr9 |
+ |
114593–114844 |
mRNA_12749 |
Hypothetical protein |
83 |
||||
|
Chr9 |
− |
115206–116625 |
mRNA_12750 |
hypothetical protein |
454 |
||||
|
Chr9 |
+ |
117388–118112 |
mRNA_12751 |
Hypothetical protein |
225 |
||||
|
Chr9 |
+ |
118451–119788 |
mRNA_12752 |
Hypothetical protein |
445 |
||||
|
Chr9 |
− |
120249–121379 |
mRNA_12753 |
Methyltransf-18 multi-domain protein |
376 |
||||
|
Chr9 |
+ |
125936–126175 |
mRNA_12755 |
Hypothetical protein |
60 |
||||
|
Chr9 |
− |
127211–127757 |
mRNA_12756 |
Hypothetical protein |
159 |
||||
|
Chr9 |
+ |
128397–131000 |
mRNA_12757 |
Cwf-Cwc-15 domain-containing protein |
867 |
||||
|
Chr9 |
− |
131128–131483 |
mRNA_12758 |
Hypothetical protein |
99 |
||||
|
Chr9 |
− |
134706–135131 |
mRNA_12759 |
Hypothetical protein |
91 |
||||
|
Chr9 |
− |
137602–138426 |
mRNA_12761 |
Hypothetical protein |
274 |
||||
|
Chr10 |
− |
2199594–2200463 |
mRNA_14375 |
Hypothetical protein |
265 |
||||
|
Chr10 |
+ |
4238821–4240083 |
mRNA_15173 |
Hypothetical protein |
421 |
PHI-base phenotype classifications: *reduced virulence, †lethal. ‡M4 mRNA in planta. §M4 mRNA in vitro.
The genes specific for ToxC-producing isolates (that were PacBio-sequenced) were also searched in previous published in planta (3 and 4 days post-inoculation) and in vitro (7- and 9-day-old vegetative and sporulating mycelia, respectively) RNA-seq data [40]. Most of the gene cluster (KAF7566087–KAF7566105) had in vitro transcription support (Fig. S13). Only the hypothetical transmembrane protein (KAF7575752) and two other hypothetical proteins had in planta transcription support during Ptr infection (Table 3).
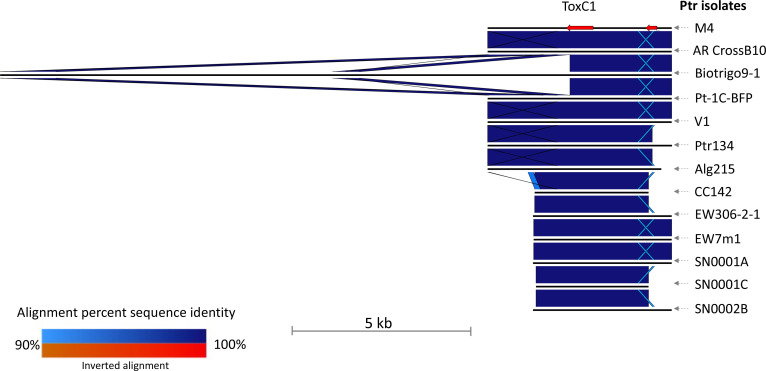
When all sequenced isolates were considered, only a single locus for a transmembrane protein, an integral membrane component, was identified core to all ToxC-producing isolates, represented by the M4 protein (KAF7575752) on chr2 position 5 052 985–5 053 659 bp (Fig. 1d). This gene was recently identified as ToxC1, a gene required but not sufficient for ToxC production in Ptr [78]. A less stringent search for ToxC1 in all isolates detected the presence of ToxC1 in the race 2 isolate Biotrigo9-1 genome, which was disrupted by a large insertion of 5 348 bp, positioned at 45 946–51 292 bp on contig 12, which disrupted the ToxC1 protein coding region in the 582–583 bp position. Examination of the 2 kb gene flanking regions of all genomes indicated a further large insertion downstream of the gene in Biotrigo9-1 (Fig. 3). The two large insertions do not have a similar sequence identity, with the insertion downstream of ToxC1 carrying Gypsy retrotransposon transposable elements (TEs) and the ToxC1 insertion carrying Copia retrotransposon TEs informed by flanking long terminal repeats (LTRs) (Fig. S14).
Fig. 3.
Ptr isolate M4 ToxC1 locus and 2 kb flanking sequence region alignment to 12 other Ptr ToxC-producing isolates. The Biotrigo9-1 ToxC1 region has two large insertions within and downstream of ToxC1. Nucleotide sequence alignments (blue) between the ToxC1 region for Ptr isolates (top to bottom: M4, AR CrossB10, Biotrigo9-1, Pt-1C-BFP, V1, Ptr134, Alg215, CC142, EW306-2-1, EW7m1, SN0001A, SN0001C and SN0002B) (black lines). The M4 genes are shown as red arrows. The light blue alignment segments are regions of low identity among the isolates, while the crossed regions indicate a repeat region in each sequence.
Analyses of core and accessory gene sets/protein clusters
To determine core and accessory protein groups in Ptr, a total of 33 1644 predicted protein coding genes from this study and published genomes downloaded from NCBI (see Methods) were clustered. Of the total number, 328 336 proteins clustered into 14 833 orthologous groups and 3 308 singletons, representing a pangenome for Ptr. A total of 8503 groups were core (57 %) (with all isolates present) and 7 162 orthogroups (48 %) consisted entirely of single-copy genes (Supplementary data 4). Overall, for the PacBio-sequenced isolates, race 4 isolate 90-2 had the highest percentage of duplicated genes (two copies) (12 %) (Supplementary data 4). The percentage of single-copy genes for the PacBio-sequenced genomes ranged from 76 % for M4 to 89 % for Ls13-192. The majority of singletons were hypothetical proteins and of those assigned a GO term the majority were related to protein binding (364 genes) (Fig. S15).
Across the Ptr pangenome (core and accessory genes), 32 257 (9.6 %) genes had a signal peptide of which just over one-third (11 911 genes) were predicted to be effectors (EffectorP 3.0 default probability score ≥0.5). The EffectorP 3.0 NE probability scores for ToxA and ToxB were 0.702 and 0.93, respectively. The NE ToxA and ToxB/toxb were identified in orthologous protein groups OG0011421 and OG0011851, respectively.
All predicted effectors protein sequences were then clustered into 738 orthogroups, of which five groups were isolate-specific (containing paralogous genes) and 187 were singletons (a single gene). Of the 738 effector orthogroups, only 119 (16 %) were core to all isolates and of the core orthogroups 25 (21 %) had 100 % sequence identity. Of the non-core effector groups, 62 orthogroups were absent in the race 4 isolates T205, Ls13-192, 90-2 and SD20.
A comparison of predicted effectors from orthogroups with race 4 absent to those with race 4 present found that the average protein length was shorter (t-test, Wilcoxon adj. P=2.9e-294) and the effector probability scores were higher (t-test, Wilcoxon adj. P=1.8–28) (Fig. S16).
Protein tertiary structure analysis of predicted effectors
To identify protein tertiary structure homology, predicted effectors were screened using remote homology detection methods against known protein structures to build 3D models. Of these, 147 proteins had predicted high-confidence tertiary models based on published tertiary protein structures (Phyre2 confidence ≥90 % and alignment coverage ≥90 %) (Supplementary data 5). Of the high-confidence proteins, 48 and 19 had annotated hydrolase and binding functions, respectively. Five were annotated as effectors, which included Ptr ToxA NE (KAF7569451) with 100 % sequence identity to the Protein Database (PDB) crystal protein structure of ToxA 1ZLD and four elicitor proteins, hrip2 (KAF7578077, KAF7575054, KAF7570798 and KAF757229), based on the crystal structure from Magnaphorthe oryzae (PDB 5FID) with sequence identities ranging between 23 and 26 %.
The 147 predicted effector proteins with a confident protein tertiary model were then searched against Phi-Base [68]. A total of 34 proteins had known Phi-Base pathogenicity or reduced virulence hits, of which 11 were plant avirulence determinants, which included ToxA (Supplementary data 5).
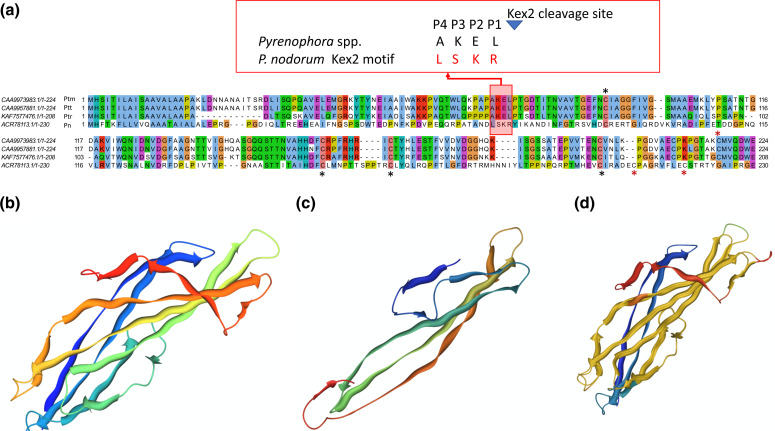
To enable the capture of genes that may have been filtered out previously (that may not have a predicted signal peptide), whole genome HMM libraries of M4 were generated for screening using BackPhyre [66]. NE-related protein structures were then selected from toxins available in the RCSB PDB for Ptr ToxB (2MM0), toxb (2MM2), ToxA (1ZLD) and SnTox3 (6WES) to identify any other structural homologues and orthologues, respectively. No structural paralogues for ToxA or ToxB were identified in isolate M4 (with confidence level ≥20.0); however, an orthologous structure was identified for SnTox3 with 58 % alignment coverage (46–138 aa) to M4 (protein accession KAF7577476) (104–195 aa in the alignment) with a confidence score of 95.5 and 34% protein sequence identity (Fig. 4a). This indicated a high confidence that the match between KAF7577476 and the PnTox3 template is a true homology that adopts the overall protein fold and that the core protein is modelled at a high accuracy (2–4 Å from the native, true structure). The 3D protein structures for SnTox3 (Fig. 4b) and predicted structure for KAF7577476 (Fig. 4c) were then structurally aligned and superimposed with a root mean square distance (RMSD) of 1.14 Å (Fig. 4d).
Fig. 4.
The predicted protein sequence and structural alignments of SnTox3 and the isolate M4 protein KAF7577476. (a) Multiple protein sequence alignment of SnTox3, Ptt CAA9973983.1 (W1-1), Ptm CAA9957881.1 (SG1) and Ptr KAF7577476 (M4). The Kex2 motif conservation is shown boxed in red. Only four cysteine residues were conserved across the four species (black asterisks) and those not conserved (red asterisks) are shown below the alignment for P. nodorum and above for the Pyrenophora species. (b) The known 3D protein structure for SnTox3 (PDB 6WES). (c) The 3D structure for KAF7577476 as predicted by Phyre2. (d) Superimposed structural alignment (yellow) of SnTox3 and KAF7577476 with an RSDM of 1.14 Å.
A TBLASTN sequence search of the M4 isolate predicted protein KAF7577476 against all the Ptr genomes found evidence that the gene encoding this predicted protein is present in all isolates. The KAF7577476 protein sequence was then also searched against the genomes of the related barley necrotrophic fungal pathogens P. teres f. teres isolate W1-1 (Ptt) and P. teres f. maculata isolate SG1 (Ptm) [51] and high-identity orthologues were also identified: CAA9973983.1 (isolate W1-1) and CAA9957881.1 (isolate SG1), respectively (Fig. 4a). An automated and combinative method for ranking top candidate effector proteins (Predector) [49] ranked Ptr KAF7577476 in the 262th position, Ptt CAA9973983 as the top candidate (number 1) and Ptm CAA9957881 in the 56th position with Predector scores of 1.9, 3.9 and 2.7, respectively (Supplementary data 1). The SnKex2 cleavage motif LSKR (69–72 aa) of SnTox3 [79] aligned to AKEL protein residues in the three Pyrenophora species (Ptr, Ptm and Ptt), where the residue positioned before the cleavage site (P1) is expected to be exclusively an arginine (Arg, R) [79] (Fig. 4a). Furthermore, the Pyrenophora sequences appeared to possess only four of the six cysteine residues, which form three disulphide bonds [80], conserved with SnTox3. The predicted apoplastic effector scores for SnTox3, KAF7577476 (Ptr), CAA9973983 (Ptt) and CAA9957881 (Ptm) were 0.573, 0.572, 0.691 and 0.765, respectively.
Discussion
P. tritici-repentis pangenome analysis
In this study, we present the pangenome of 26 Ptr isolates with a near complete representation of the eight known race categories. Our 15 newly assembled and annotated genomes, along with the 11 previously published genomes, represent a global pangenome of Ptr for major wheat-growing regions, with close and distant proximity to the origin of wheat domestication in the Fertile Crescent of western Asia. The repertoire of the known Ptr genes [14] was expanded by 31 %, represented by 18 140 non-redundant sequences. This expansion of genes is also observed in other plant fungal species, where a pangenome analysis of 20 F. graminearum isolates resulted in a 32 % gene expansion over the reference isolate [20]. The 57 % conservation of core orthogroups in Ptr identified here is similar in magnitude to a recent 19 isolate pangenome analysis of the wheat pathogen Z. tritici [21], which found that 60 % of gene orthogroups were core.
A number of Ascomycete genomes, such as Pn, Ptt and Zt, have ‘two speed genomes’, where the genome is compartmentalized into gene-poor AT-rich regions and can have accessory chromosomes. In contrast, Ptr does not appear to have accessory chromosomes and has a GC-equilibrated genome [14, 51, 81–83]. Whole genome phylogenetic analysis clearly showed greater isolate phylogenetic distances within Bs, Pn and Zt isolates as compared with the Pyrenophora species (Ptm, Ptr and Ptt). However, even with comparatively low phylogenetic distances within Ptr, distinct clades could be detected based on geographical locations. The only exception was isolate Alg215 from Algeria, which clustered with the Australian and American isolates, sharing a large sub-telomeric region in common. This sequence variation, plus a disrupted ToxB, set Alg215 apart from the other isolates collected from North Africa. Despite the low whole-genome phylogenetic distances in Ptr, a lower percentage of core orthogroups (57 %) was found compared with a previous analyses of 11 isolates (PacBio- and Illumina-sequenced), which indicated 69 % core orthologous groups [14]. This suggests that not only has the pangenome complexity risen with an increase in the numbers of isolates sequenced (as expected), but that an increased divergence in Ptr conserved protein domains is apparent.
Although in this analysis only a single gene was identified as specific to all ToxC-producing isolates (ToxC1), PacBio sequencing identified a potential gene cluster of interest which would be near impossible to identify in Illumina-sequenced genomes, due to the repetitive nature of the region. Interestingly, our analysis found no putative effectors that were core to all isolates, again indicating a large variability within Ptr for this type of gene. Recently, ToxC1 was functionally validated using a gene knockout approach [78], where it was found to be required, but not sufficient, for ToxC production. In our study, no clear gene cluster for a secondary metabolite or ribosomally synthesized and post-translationally modified peptides (RiPPs) was identified, in part due to the positioning of the ToxC locus within the complex subtelomeric region of chromosome 2 [78], which despite long read sequencing still remains a problematic region to resolve. The presence of ToxC1 in a non-ToxC-producing isolate (Biotrigo9-1) was surprising, and raises more questions regarding the evolution and/or origin of ToxC production. It is possible that the large ToxC1 insertion by an LTR retrotransposon has disrupted the production of ToxC in Biotrigo9-1 and that those remaining gene(s) involved in ToxC production are present.
The divergence of Ptr race 4 isolates (that do not produce the known NEs on wheat) from isolates that produce known NEs was clearly shown, except for T205. Given the same assembly methods were used for all the PacBio genomes, the genome sizes and gene duplication rates of the two race 4 isolates (Ls13-192 and 90-2) revealed a complexity that was unexpected. Race 4 was first described 30 years ago [72, 84] as a nec− chl− pathotype (avirulent) on the set of differential wheat lines, and has since been reported but not as frequently as the other races from collections of Ptr across different wheat-growing regions. A recent study [69] showed that despite the inability of race 4 isolates to induce tan spot symptoms on the differential wheat lines, four race 4 isolates (Ls13-14, Ls13-86, Ls13-192 and Ls13-198) from North Dakota in the USA induced varying degrees of disease reactions upon inoculation on tetraploid (durum) wheats [69]. This may well provide an explanation for the observed distinction of Ls13-192 from the other race 4 isolates (90-2 and SD20) in the whole genome phylogenetic clustering and gene correlation analyses, since unlike Ls13-192, SD20 and 90-2 have not been reported to be virulent on durum wheat. Furthermore, as ToxA and ToxB were absent, race 4 isolate T205 is unlike the new virulence type that lacked ToxA and ToxB gene expression on bread wheat differentials but produced necrosis in durum wheat [85].
While it was unexpected that a race 4 isolate (90-2) had the highest percentage of genes predicted as effectors, this appeared to be the result of a genome-wide expansion of gene copies (which included predicted effectors). It is possible that although the predicted effectors in race 4 isolates may have not have a pathogenic role in bread wheat, they may play a role in another system.
We report here, for the first time, an identical toxb (non-toxic homologue of ToxB) copy in a race 4 genome (two genes in 90-2). As each toxb is on separate contigs, it is not possible to identify if they are co-located. We can, however, speculate that based on the difficulty in assembling the toxb regions, they may lie in a subtelomeric chromosome location similar to the multicopy ToxB, which was shown to be nested in the complex subtelomeric chromosomal regions of the DW5 genome [73]. It is increasingly believed that effector genes are located in transposon-rich and gene-sparse subtelomeric regions of the pathogen genome, allowing opportunity for gene duplication events and thereby contributing to the evolution of virulence diversity. We also show no conservation between the different races for the ToxB locus or flanking regions. The sequence variation in the chromosomal centromeric and telomeric regions shown in our whole genome alignments indicates that these regions are indeed hotspots for diversity. It is furthermore interesting that one of the North Africa isolates (Alg 215) had a truncated ToxB gene with a non-synonymous mutation within the coding region, which may have resulted in a weak chlorosis phenotype on the wheat differential lines. We believe that this is the first report of a ToxB non-synonymous mutation in Ptr. The large ToxA horizontal transfer region previously identified [14, 23] was shown to be absent in all non-ToxA-producing isolates, and clear insertion breakpoints were identified in all ToxA-producing isolates.
In this study, a pangenome approach was undertaken to approximate the complete gene repertoire of the species to capture all gene variations (percentage identity and copy number) and identify candidate genes specific for ToxC-producing races.
A new Pyrenophora resource to identify protein structural homologues
Pathogenic fungi possess large effector repertoires that are dominated by hundreds of small secreted proteins only related by protein tertiary structures (3D structure) [19]. The prediction of new effector candidates that are not the result of horizontal gene transfer is therefore complicated.
To conduct a comprehensive whole genome search of protein tertiary structures an in silico screening was employed using BackPhyre [66]. We present here the first necrotrophic fungal pathogen publicly available through BackPhyre [66] for effector and other protein tertiary structure searches, providing further annotation evidence for a number of hypothetical genes. In this pangenome screen of proteins, no other ToxA or ToxB-like paralogues were identified based on structural similarity in Ptr.
Overall, the use of protein 3D structure modelling improved the identification of a number of proteins which included effector candidates potentially involved in pathogenicity.
In silico protein structural analysis reveals a natural homologue to SnTox3 in Pyrenophora
We report here for the first time a distant SnTox3 natural homologue in Pyrenophora. We showed conserved structural homology between SnTox3 and Pyrenophora proteins that lacked conservation in the R residue position of the Kex2 motif (LXXR) and the full set of cysteine residues forming the three disulphide bonds in SnTox3. SnTox3 is a pro-domain-containing NE, where the signal peptide and pro-domain are removed (cleaved by the Kex2 protease) to produce a more potent protein that activates host cell death (Snn3) [86]. The Kex2 cleavage motif (LXXR) has the following residue preferences, a leucine (L, Leu) or any other aliphatic residue, any residue X as it does not interact with Kex2, lysine (K, Lys) but has other possible residues Lys>arginine (R, Arg)>threonine (T, Thr)>proline (P, Pro)>glutamic acid (E, Glu)>isoleucine (I, Ile) (X) and exclusively an arginine (R, Arg) before the cleavage site [79]. Here we found the conserved Pyrenophora motif (AKEL) did not conform to the Kex2 cleavage motif (LXXR) in two residue positions that included the exclusive arginine residue.
Interestingly, the Ptr structural homologue to SnTox3 was in all isolate races, unlike the non-active toxb that only occurs in non-pathogenic race 4 isolates (not producing known NEs). As no in planta gene expression for the Ptr homologue of SnTox3 was detected and the protein sequence had a low effector prediction ranking, we believe it may not be an effector candidate in the wheat–pathogen system. However, conversely in Ptt, as the structural homologue to SnTox3 is expressed during barley infection [87] and was ranked as the top candidate effector, we believe further investigation is warranted. Here, we propose that the identification of an SnTox3 structural homologue in Pyrenophora (Ptm, Ptr and Ptt) could be part of a structurally defined family that are phylogenetically related to SnTox3, as observed for the M. oryzae Avirulence (Avrs) and ToxB (MAX-effector proteins) [19].
In conclusion, the new genomic resources presented here improve the pangenome representation of Ptr and provide putative effector candidates based on structural modelling and ranking specific to effector-producing isolates. These resources can be used to monitor Ptr variations potentially involved in pathogenicity. As Ptr is commonly shown to infect wheat in combination with other necrotrophic pathogens [88], the future ability to simultaneously monitor such changes in multiple necrotrophic species may enhance pathogen monitoring activities within a wider framework of crop protection activities.
Supplementary Data
Funding information
This work was generously supported through co-investment by the Grains Research and Development Corporation (GRDC) and Curtin University (project code CUR00023), as well as the Australian Government National Collaborative Research Infrastructure Strategy and Education Investment Fund Super Science Initiative. This project was also supported by the Agriculture and Food Research Initiative competitive grants programme (award number 2016-67014-24806) and the National Institute of Food and Agriculture, United States Department of Agriculture (USDA) Hatch project (ND02234) to Z.L. J.C., J.T. and L.J. were supported by the ‘Efectawheat’ project funded within the framework of the 2nd call ERA-NET for Coordinating Plant Sciences by the British Biological Sciences Research Council (BBSRC) grant BB/N00518X/1 to J.C. and the Danish Council of Strategic Research grant case number 5147-00002B to L.J. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgements
We thank the Australian grain growers for their continued support of research through the Grains Research and Development Corporation (GRDC) and the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS) for providing access to Pawsey Supercomputing under a National Computational Merit Allocation Scheme (NCMAS), Nectar Research and Pawsey Nimbus Cloud resources. We would also like to acknowledge Professor Richard Oliver who was key in the setting up the collaborators in this study as part of the EfectaWheat project.
Author contributions
Conceptualization Z.L. and C.S.M.; methodology, P.M., P.T.S. and H.P.; formal analysis, P.M., P.T.S., G.S. and H.P.; investigation, P.M.; project resources J.C., J.T., S.S., H.B. and L.J.; writing - original draft preparation, P.M.; writing – review and editing, P.M., P.T.S., Z.L., G.S., H.B., J.C., L.J., S.S. and C.M. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare no competing interests.
Footnotes
Abbreviations: CDS, coding sequence; chr, chromosome; HMM, Hidden Markov Model; LTR, long terminal repeat; NE, necrotrophic effector; NRPS, non-ribosomal protein synthase; Ptr, Pyrenophora tritici-repentis; RMSD, root mean square distance; Sn, Parastagonospora nodrum; TE, transposable element; Zt, Zymoseptoria tritici.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary data sets and sixteen supplementary figures are available with the online version of this article.
References
- 1.Benslimane H, Lamari L, Benbelkacem A, Sayoud R, Bouznad Z. Distribution of races of Pyrenophora tritici-repentis in Algeria and identication of a new virulence type. Phytopathol Mediterr. 2011;50:203–211. [Google Scholar]
- 2.Murray GM, Brennan JP. Estimating disease losses to the Australian wheat industry. Austral Plant Pathol. 2009;38:558. doi: 10.1071/AP09053. [DOI] [Google Scholar]
- 3.Bhathal JS, Loughman R, Speijers J. Yield reduction in wheat in relation to leaf disease from yellow (tan) spot and Septoria nodorum blotch. Eur J Plant Pathol. 2003;109:435–443. doi: 10.1023/A:1024277420773. [DOI] [Google Scholar]
- 4.Ciuffetti LM, Manning VA, Pandelova I, Betts MF, Martinez JP. Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol. 2010;187:911–919. doi: 10.1111/j.1469-8137.2010.03362.x. [DOI] [PubMed] [Google Scholar]
- 5.Downie RC, Lin M, Corsi B, Ficke A, Lillemo M, et al. Septoria nodorum blotch of wheat: disease management and resistance breeding in the face of shifting disease dynamics and a changing environment. Phytopathology. 2021;111:906–920. doi: 10.1094/PHYTO-07-20-0280-RVW. [DOI] [PubMed] [Google Scholar]
- 6.Faris JD, Liu Z, Xu SS. Genetics of tan spot resistance in wheat. Theor Appl Genet. 2013;126:2197–2217. doi: 10.1007/s00122-013-2157-y. [DOI] [PubMed] [Google Scholar]
- 7.Friesen TL, Zhang Z, Solomon PS, Oliver RP, Faris JD. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 2008;146:682–693. doi: 10.1104/pp.107.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciuffetti LM, Tuori RP, Gaventa JM. A single gene encodes selective toxin causal to the development of tan spot of wheat. Plant Cell. 1997;9:135–144. doi: 10.1105/tpc.9.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strelkov SE, Lamari L, Ballance GM. Characterization of a host-specific protein Toxin (Ptr ToxB) from Pyrenophora tritici-repentis . MPMI. 1999;12:728–732. doi: 10.1094/MPMI.1999.12.8.728. [DOI] [Google Scholar]
- 10.Lamari L, Strelkov SE. The wheat/pyrenophora tritici-repentis interaction: progress towards an understanding of tan spot disease. Can J Plant Pathol. 2010;32:4–10. [Google Scholar]
- 11.Ali S, Gurung S, Adhikari TB. Identification and characterization of novel isolates of Pyrenophora tritici-repentis from Arkansas. Plant Dis. 2010;94:229–235. doi: 10.1094/PDIS-94-2-0229. [DOI] [PubMed] [Google Scholar]
- 12.Kamel S, Cherif M, Hafez M, Despins T, Aboukhaddour R. Pyrenophora tritici-repentis in Tunisia: race structure and effector genes. Front Plant Sci. 2019;10:1562. doi: 10.3389/fpls.2019.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kariyawasam GK, Wyatt N, Shi G, Liu S, Yan C, et al. A genome-wide genetic linkage map and reference quality genome sequence for A new race in the wheat pathogen Pyrenophora tritici-repentis. Fungal Genet Biol. 2021;152:103571. doi: 10.1016/j.fgb.2021.103571. [DOI] [PubMed] [Google Scholar]
- 14.Moolhuijzen P, See PT, Hane JK, Shi G, Liu Z, et al. Comparative genomics of the wheat fungal pathogen Pyrenophora tritici-repentis reveals chromosomal variations and genome plasticity. BMC Genomics. 2018;19:279. doi: 10.1186/s12864-018-4680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rybak K, See PT, Phan HTT, Syme RA, Moffat CS, et al. A functionally conserved Zn2 Cys6 binuclear cluster transcription factor class regulates necrotrophic effector gene expression and host-specific virulence of two major Pleosporales fungal pathogens of wheat. Mol Plant Pathol. 2017;18:420–434. doi: 10.1111/mpp.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.See PT, Marathamuthu KA, Iagallo EM, Oliver RP, Moffat CS. Evaluating the importance of the tan spot ToxA- Tsn1 interaction in Australian wheat varieties. Plant Pathol. 2018;67:1066–1075. doi: 10.1111/ppa.12835. [DOI] [Google Scholar]
- 17.Andrie RM, Pandelova I, Ciuffetti LM. A combination of phenotypic and genotypic characterization strengthens Pyrenophora tritici-repentis race identification. Phytopathology. 2007;97:694–701. doi: 10.1094/PHYTO-97-6-0694. [DOI] [PubMed] [Google Scholar]
- 18.Tuori RP, Wolpert TJ, Ciuffetti LM. Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis . Mol Plant Microbe Interact. 1995;8:41–48. doi: 10.1094/mpmi-8-0041. [DOI] [PubMed] [Google Scholar]
- 19.de Guillen K, Ortiz-Vallejo D, Gracy J, Fournier E, Kroj T, et al. Structure analysis uncovers a highly diverse but structurally conserved effector family in Phytopathogenic ungi. PLoS Pathog. 2015;11:e1005228. doi: 10.1371/journal.ppat.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alouane T, Rimbert H, Bormann J, González-Montiel GA, Loesgen S, et al. Comparative genomics of eight Fusarium graminearum strains with contrasting aggressiveness reveals an expanded open pangenome and extended effector content ignatures. Int J Mol Sci. 2021;22:12. doi: 10.3390/ijms22126257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Badet T, Oggenfuss U, Abraham L, McDonald BA, Croll D. A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici . BMC Biol. 2020;18:12. doi: 10.1186/s12915-020-0744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertazzoni S, Jones DAB, Phan HT, Tan KC, Hane JK. Chromosome-level genome assembly and manually-curated proteome of model necrotroph Parastagonospora nodorum Sn15 reveals a genome-wide trove of candidate effector homologs, and redundancy of virulence-related functions within an accessory chromosome. BMC Genomics. 2021;22:382. doi: 10.1186/s12864-021-07699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning VA, Pandelova I, Dhillon B, Wilhelm LJ, Goodwin SB, et al. Comparative genomics of a plant-pathogenic fungus, Pyrenophora tritici-repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3. 2013;3:41–63. doi: 10.1534/g3.112.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards JK, Wyatt NA, Liu Z, Faris JD, Friesen TL. Reference quality genome assemblies of three Parastagonospora nodorum isolates differing in virulence on wheat. G3. 2018;8:393–399. doi: 10.1534/g3.117.300462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernikos G, Medini D, Riley DR, Tettelin H. Ten years of pan-genome analyses. Curr Opin Microbiol. 2015;23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Moolhuijzen P, See PT, Moffat CS. A new PacBio genome sequence of an Australian Pyrenophora tritici-repentis race 1 isolate. BMC Res Notes. 2019;12:642. doi: 10.1186/s13104-019-4681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffat CS, See PT, Oliver RP. Generation of a ToxA knockout strain of the wheat tan spot pathogen Pyrenophora tritici-repentis . Mol Plant Pathol. 2014;15:918–926. doi: 10.1111/mpp.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamari L, Sayoud R, Boulif M, Bernier CC. Identification of a new race in Pyrenophora tritici-repentis: implications for the current pathotype classification system. Can J Plant Pathol. 1995;17:312–318. doi: 10.1080/07060669509500668. [DOI] [Google Scholar]
- 29.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews S. “FastQC.” Retrieved 2016. 2011. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 33.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen N. Using repeatmasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2004;Chapter 4:Unit 4.10. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- 37.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borodovsky M, Lomsadze A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics. 2011;Chapter 4:Unit. doi: 10.1002/0471250953.bi0406s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Testa AC, Hane JK, Ellwood SR, Oliver RP. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics. 2015;16:170. doi: 10.1186/s12864-015-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moolhuijzen P, See PT, Moffat CS. Exploration of wheat and pathogen transcriptomes during tan spot infection. BMC Res Notes. 2018;11:907. doi: 10.1186/s13104-018-3993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiryev SA, Papadopoulos JS, Schäffer AA, Agarwala R. Improved BLAST searches using longer words for protein seeding. Bioinformatics. 2007;23:2949–2951. doi: 10.1093/bioinformatics/btm479. [DOI] [PubMed] [Google Scholar]
- 44.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 47.Sperschneider J, Dodds PN. EffectorP 3.0: prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol Plant Microbe Interact. 2021 doi: 10.1101/2021.07.28.454080. [DOI] [PubMed]
- 48.Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 2018;19:2094–2110. doi: 10.1111/mpp.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones DAB, Rozano L, Debler JW, Mancera RL, Moolhuijzen PM, et al. An automated and combinative method for the predictive ranking of candidate effector proteins of fungal plant pathogens. Sci Rep. 2021;11:19731. doi: 10.1038/s41598-021-99363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 51.Syme RA, Martin A, Wyatt NA, Lawrence JA, Muria-Gonzalez MJ, et al. Transposable element genomic fissuring in Pyrenophora teres is associated with genome expansion and dynamics of host-pathogen genetic interactions. Front Genet. 2018;9:130. doi: 10.3389/fgene.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syme RA, Tan K-C, Hane JK, Dodhia K, Stoll T, et al. Comprehensive annotation of the Parastagonospora nodorum reference genome using next-generation genomics, transcriptomics and proteogenomics. PLoS One. 2016;11:e0147221. doi: 10.1371/journal.pone.0147221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald MC, Ahren D, Simpfendorfer S, Milgate A, Solomon PS. The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana . Mol Plant Pathol. 2018;19:432–439. doi: 10.1111/mpp.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klötzl F, Haubold B. Phylonium: fast estimation of evolutionary distances from large samples of similar genomes. Bioinformatics. 2020;36:2040–2046. doi: 10.1093/bioinformatics/btz903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 56.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;Chapter 10:Unit. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 57.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Team R. C R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing. 2021.
- 59.RStudio-Team “RStudio: Integrated Development Environment for R.” 1.3.1093. 2020. http://www.rstudio.com/
- 60.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olson SA. EMBOSS opens up sequence analysis. Brief Bioinformatics. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 63.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–21. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rotkiewicz P. iMol Molecular Visualization Program. 2007. http://www.pirx.com/iMol
- 68.Urban M, Cuzick A, Rutherford K, Irvine A, Pedro H, et al. PHI-base: a new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017;45:D604–D610. doi: 10.1093/nar/gkw1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo J, Shi G, Kalil A, Friskop A, Elias E, et al. Pyrenophora tritici-repentis Race 4 isolates cause disease on tetraploid wheat. Phytopathology. 2020;110:1781–1790. doi: 10.1094/PHYTO-05-20-0179-R. [DOI] [PubMed] [Google Scholar]
- 70.Lamari L, Gilbert J, Tekauz A. Race differentiation in Pyrenophora tritici-repentis and survey of physiologic variation in western Canada. Can J Plant Pathol. 1998;20:396–400. doi: 10.1080/07060669809500410. [DOI] [Google Scholar]
- 71.Bertagnolli VV, Ferreira JR, Liu ZH, Rosa AC, Deuner CC. Phenotypical and genotypical characterization of Pyrenophora tritici-repentis races in Brazil. Eur J Plant Pathol. 2019;154:995–1007. doi: 10.1007/s10658-019-01720-3. [DOI] [Google Scholar]
- 72.Lamari L, Bernier CC. Virulence of isolates of Pyrenophora tritici-repentis on 11 wheat cultivars and cytology of the differential host reactions. Can J Plant Pathol. 1989;11:284–290. doi: 10.1080/07060668909501114. [DOI] [Google Scholar]
- 73.Moolhuijzen P, See PT, Moffat CS. PacBio genome sequencing reveals new insights into the genomic organisation of the multi-copy ToxB gene of the wheat fungal pathogen Pyrenophora tritici-repentis . BMC Genomics. 2020;21:645. doi: 10.1186/s12864-020-07029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Figueroa Betts M, Manning VA, Cardwell KB, Pandelova I, Ciuffetti LM. The importance of the N-terminus for activity of Ptr ToxB, a chlorosis-inducing host-selective toxin produced by Pyrenophora tritici-repentis . Physiological and Molecular Plant Pathology. 2011;75:138–145. doi: 10.1016/j.pmpp.2011.03.002. [DOI] [Google Scholar]
- 75.Kim YM, Strelkov SE. Heterologous expression and activity of Ptr ToxB from virulent and avirulent isolates of Pyrenophora tritici-repentis . Can J Plant Pathol. 2007;29:232–242. doi: 10.1080/07060660709507465. [DOI] [Google Scholar]
- 76.Moolhuijzen PM, See PT, Oliver RP, Moffat CS. Genomic distribution of a novel Pyrenophora tritici-repentis ToxA insertion element. PLoS One. 2018;13:e0206586. doi: 10.1371/journal.pone.0206586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 2006;38:953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- 78.Shi G, Kariyawasam G, Liu S, Leng Y, Zhong S, et al. A conserved hypothetical gene is required but not sufficient for Ptr ToxC production in Pyrenophora tritici-repentis . Mol Plant Microbe Interact. 2022;35:336–348. doi: 10.1094/MPMI-12-21-0299-R. [DOI] [PubMed] [Google Scholar]
- 79.Outram MA, Solomon PS, Williams SJ. Pro-domain processing of fungal effector proteins from plant pathogens. PLoS Pathog. 2021;17:e1010000. doi: 10.1371/journal.ppat.1010000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osbourn A. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 81.Bertazzoni S, Williams AH, Jones DA, Syme RA, Tan K-C, et al. Accessories make the outfit: accessory chromosomes and other dispensable DNA regions in plant-pathogenic ungi. Mol Plant Microbe Interact. 2018;31:779–788. doi: 10.1094/MPMI-06-17-0135-FI. [DOI] [PubMed] [Google Scholar]
- 82.Dong S, Raffaele S, Kamoun S. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev. 2015;35:57–65. doi: 10.1016/j.gde.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 83.Testa AC, Oliver RP, Hane JK. OcculterCut: A Comprehensive Survey of AT-Rich Regions in Fungal Genomes. Genome Biol Evol. 2016;8:2044–2064. doi: 10.1093/gbe/evw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lamari L, Bernier CC. Genetics of tan necrosis and extensive chlorosis in tan spot of wheat caused by Pyrenophora tritici-repentis . Phytopathology. 1991;81:1092. doi: 10.1094/Phyto-81-1092. [DOI] [Google Scholar]
- 85.Benslimane H. Virulence phenotyping and molecular characterization of a new virulence type of Pyrenophora tritici-repentis the causal agent of tan spot. Plant Pathol J. 2018;34:139–142. doi: 10.5423/PPJ.NT.07.2017.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Outram MA, Sung Y-C, Yu D, Dagvadorj B, Rima SA, et al. The crystal structure of SnTox3 from the necrotrophic fungus Parastagonospora nodorum reveals a unique effector fold and provides insight into Snn3 recognition and pro-domain protease processing of fungal effectors. New Phytol. 2021;231:2282–2296. doi: 10.1111/nph.17516. [DOI] [PubMed] [Google Scholar]
- 87.Moolhuijzen P, Lawrence JA, Ellwood SR. Potentiators of disease During Barley Infection by Pyrenophora teres f. teres in a Susceptible Interaction. MPMI. 2021;34:779–792. doi: 10.1094/MPMI-10-20-0297-R. [DOI] [PubMed] [Google Scholar]
- 88.Justesen AF, Corsi B, Ficke A, Hartl L, Holdgate S, et al. Hidden in plain sight: a molecular field survey of three wheat leaf blotch fungal diseases in North-Western Europe shows co-infection is widespread. Eur J Plant Pathol. 2021;160:949–962. doi: 10.1007/s10658-021-02298-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.