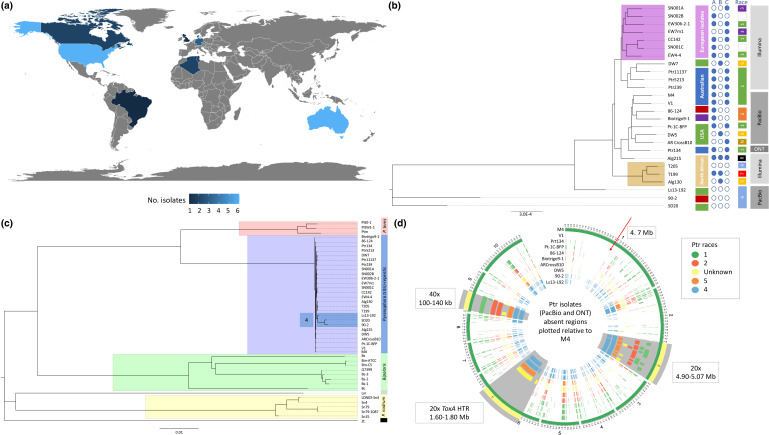
Fig. 1.
Whole genome analysis of Ptr isolates. (a) Geographical source and number of Ptr isolates currently available and analysed. Key gives the number of isolates. (b) Whole genome phylogenetic tree of Ptr isolates from Illumina sequencing (Alg130, T199, T205, Alg215, CC142, EW306-2-1, EW4-4, EW7m1, DW7, Pt-1C-BFP, Ptr239, Ptr11137, Ptr5213, SD20, SN001A, SN001C, SN002B), PacBio Technologies (86-124, 90-2, AR CrossB10, Biotrigo9-1, DW5, Ls13-192, M4 and V1) and Oxford Nanopore Technologies (Ptr134). The unrooted neighbour-joining phylogenetic tree displays clades for the European (violet) and North Africa (tan) isolates. Geographical source of the other isolates is Australia (blue), USA (green), Canada (red) and Brazil (purple). The race 4 isolates (Ls13-192, 90-2 and SD20) have the greatest distance from the clade of known NE-producing isolates. (c) Unrooted neighbour-joining phylogenetic tree for Ptr (purple clade), Pyrenophora teres [P. teres f. maculata (Ptm) and P. teres f. teres (Ptt)) (orange clade), Bipolaris [B. sorokiniana (Bs1-3 and Q7399), B. maydis (Bm-ATCC and Bm-C5) and Bipolaris zeicola (Bz)] (green clade), Parastagonospora nodorum (Sn4, Sn15 and Sn79) (yellow clade), Leptosphaeria maculans (Lm) and Zymoseptoria tritici (Zt) isolates. The branches for race 4 isolates not producing known NEs (Ls13-192, 90-2 and SD20) are highlighted (blue) within the Ptr clade. (d) Circular plots show 10 kb regions of absence plotted for the Ptr isolate genomes sequenced using long-read technologies (PacBio and Oxford Nanopore Technology) as compared with the chromosomes of the reference Ptr genome of isolate M4. Isolates are coloured by race. Three regions of interest are highlighted in grey and zoomed at 20× for chromosome 2 and chromosome 1, and 40× for chromosome 9.