Abstract
The cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) signaling pathway plays a vital role in sensing viral DNA in the cytosol, stimulating type I interferon (IFN) production and triggering the innate immune response against DNA virus infection. However, viruses have evolved effective inhibitors to impede this sensing pathway. Chicken anemia virus (CAV), a nonenveloped ssDNA virus, is a ubiquitous pathogen causing great economic losses to the poultry industry globally. CAV infection is reported to downregulate type I IFN induction. However, whether the cGAS-STING signal axis is used by CAV to regulate type I IFN remains unclear. Our results demonstrate that CAV infection significantly elevates the expression of cGAS and STING at the mRNA level, whereas IFN-β levels are reduced. Furthermore, IFN-β activation was completely blocked by the structural protein VP1 of CAV in interferon stimulatory DNA (ISD) or STING-stimulated cells. VP1 was further confirmed as an inhibitor by interacting with interferon regulatory factor 7 (IRF7) by binding its C-terminal 143–492 aa region. IRF7 dimerization induced by TANK binding kinase 1 (TBK1) could be inhibited by VP1 in a dose-dependent manner. Together, our study demonstrates that CAV VP1 is an effective inhibitor that interacts with IRF7 and antagonizes cGAS-STING pathway-mediated IFN-β activation. These findings reveal a new mechanism of immune evasion by CAV.
Key words: chicken anemia virus, type I interferon, VP1, interferon regulatory factor 7, dimerization
INTRODUCTION
The innate immune system through germline-encoded pattern recognition receptors (PRRs) recognizes evolutionarily conserved pathogen-associated molecular patterns (PAMPs) (Ishii et al., 2006), and then initializes the host immune response against pathogen infection. Type I interferon (IFN) plays an essential part in the innate immune responses against viruses. The cyclic GMP-AMP synthase (cGAS), one of the predominant cytosolic DNA sensors of PRRs, is critical in the activation of the type I IFN expression. Upon sensing cytoplasmic “danger” signals such as DNA or RNA derived from a large variety of DNA-containing pathogens, cGAS synthesizes cyclic GMP-AMP and then activates the stimulator of interferon genes (STING), also known as TMEM173, MITA, ERIS, or MPYS (Ishikawa et al., 2009; Sun et al., 2013; Motwani et al., 2021). STING activation usually results in the recruitment and phosphorylation of adaptor molecules, such as TANK binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3). Phosphorylated IRF3 then dimerizes and induces the secretion of type I IFN and a series of other downstream cytokines, which plays a major role in innate defense against DNA virus infection (Wu et al., 2013; Shu et al., 2014; Xia et al., 2016). Lack of the cGAS-STING sensing signal pathway remarkably reduces the host's innate immune response against DNA virus infection (Ishikawa et al., 2009; Ablasser et al., 2013; Thomsen et al., 2016). Therefore, cGAS, STING, and other signal adaptor molecules are considered attractive targets to identify novel biomarkers.
However, viruses have evolved escape mechanisms to suppress the cGAS-STING pathway driving type I interferons via viral distinct inhibitors. The mechanisms used by viral proteins to inhibit the cGAS-STING pathway are varied. One such example is herpes simplex virus 1 (HSV-1), a highly contagious infectious DNA virus, which selectively blocks IRF3 expression by viral VP24 protein, thereby inhibiting IFN expression (Zhang et al., 2016). African swine fever virus causes a severe animal infectious disease and was identified as a strong inhibitory effector of IFN β production via a viral E2 ubiquitin-conjugating enzyme, which regulates through recruiting pI215L-binding RING finger protein 138 to inhibit K63-linked ubiquitination of TBK1 (Huang et al., 2021). Human cytomegalovirus protein UL82 was also shown to bind to STING, which is blocked from activating TBK1 (Fu et al.. 2017).
Chicken anemia virus (CAV), a nonenveloped small ssDNA virus, is a ubiquitous avian pathogen that causes an immunosuppressive infectious anemia disease. This virus represses the immune responses of the host and thus, infected chickens are more susceptible to other pathogens (Wani et al., 2006; Schat, 2009; Dong et al., 2022). As a result, great economic loss has been caused by CAV in the poultry industry globally. A previous report showed that CAV infection efficiently blocks chicken type I IFN production and the subsequent cascades as well (Giotis et al., 2018). However, the underlying molecular mechanisms remain elusive. CAV has a small genome that encodes three proteins. VP1, a 52 kDa protein, is the sole structural protein for virus encapsulation (Schat, 2009). In this study, we show that CAV protein VP1 plays an essential role in regulating the cGAS-STING signal axis-induced expression of IFN-β and, more specifically, adaptor IRF7. As chickens lack classical IRF3, IRF7 may serve as a new vital target to manipulate CAV immune escape.
MATERIALS AND METHODS
Cells, Virus, and Reagents
The chicken lymphoblastoid cell line MDCC-MSB1 (MSB1) originally gifted by Dr. D. Yin (Anhui Academy of Agricultural Sciences, Hefei, China) was cultured in RPMI-1640 (Thermo Fisher Scientific, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, TransGen Biotech, Beijing, China), 10% tryptone phosphate broth (TPB, Sigma-Aldrich, Darmstadt, Germany) and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) (TransGen Biotech, Beijing, China) in an incubator at 37°C in 5% CO2. The chicken embryo fibroblast cell line (DF-1 cells), chicken macrophage cell line HD11, and HEK293T cells were originally provided by Dr. D. Yin and maintained in DMEM supplemented with 10% FBS and antibiotics. CAV strain AHAV-CAV2 (GenBank accession number MH801130) was isolated in China in 2019 and maintained in our laboratory. The antibodies used were mouse anti-mCherry, mouse anti-Myc, mouse anti-GFP, mouse anti-GAPDH, goat anti-mouse, and goat anti-rabbit HRP-IgG (Enogene Biotech, Nanjing, China). Rabbit anti-TBK1, anti-CAV-VP1, anti-cGAS, anti-STING, and anti-IRF7 polyclonal antibodies were prepared in our laboratory according to a methodology similar to that described in a previous paper (Liu et al., 2016). All animal experiments were performed using female New Zealand white rabbits around 1.5 kg purchased from the experimental animal center of Anhui Medical University. All animal procedures were conducted in accordance with the Animal Ethics Committee of Anhui Agricultural University (Approval No. SYXK2016-007) and all efforts were made to minimize suffering.
Expression Constructs
For VP1 constructs, the VP1 gene generated from MSB1 cells-derived cDNAs post-CAV infection was cloned into pCMV-Myc, pmCherry-C1, or pEGFP-C1 vectors using PCR amplification and standard cloning techniques, respectively.
For chicken cGAS (GenBank accession number XM_419881), STING (GenBank accession number KP_893157), TBK1 (GenBank accession number NM_001199558), and IRF7 (GenBank accession number KP_096419) expression, cDNA encoding the indicated protein was inserted into the pCMV-Myc or pEGFP-C1 vectors using specific primers designed and listed in Table 1.
Table 1.
Oligonucleotide sequences for PCR and luciferase reporter assays.
| Gene | Name | Oligonucleotide sequences (5′-3′) |
|---|---|---|
| cGAS | Myc-cGAS-F | GGAATTCGGATGGAGGAGACCGCGGCGGGCA |
| Myc-cGAS-R | GGGGTACCCTACACCTGGTGAAATACTGGGAATCC | |
| CAV-VP1 | mCherry-VP1-F | CCGCTCGAGAGATGGCAAGACGAGCTCGCAGAC |
| mCherry-VP1-R | GGAATTCAATCAGGGCTGCGTCCCCCAG | |
| EGFP-VP1-F | GGAATTCTATGGCAAGACGAGCTCGCAGAC | |
| EGFP-VP1-R | GGGGTACCTCAGGGCTGCGTCCCCCAG | |
| STING | Myc-STING-F | GGAATTCGGATGCCCCAGGACCCGTCAACC |
| Myc-STING-R | GGGGTACCTCAGGGGCAGTCACTGCGCAG | |
| IRF7 | Myc-IRF7-F | GGAATTCGGATGGCAGCACTGGACAGCG |
| Myc-IRF7-R | GGGGTACCTCAGTCTGTCTGCATGTGGTATTG | |
| EGEP-IRF7-F | GGAATTCTATGGCAGCACTGGACAGCG | |
| EGEP-IRF7-R | GGGGTACCTCAGTCTGTCTGCATGTGGTATTG | |
| cGAS | cGAS-luc-F | CCGCTCGAGGAAAAGAGTCACCACAAAAATCTGGC |
| cGAS-luc-R | CCCAAGCTTCGCCTCTCGGGCGGCACA | |
| STING | STING-luc-F | GGGGTACCTGAACAGGGTGGGGGTACTGCTGGA |
| STING-luc-R | CCGCTCGAGTGCCCTGTGCCTGCCTCCTGTCTG | |
| TBK1 | TBK1-luc-F | GGGGTACCCTTGGAGTCCTGCTGGTCCTCTTC |
| TBK1-luc-R | CCCAAGCTTGAGCAGCGCGCGGCGCG | |
| IRF7 | IRF3-luc-F | GGGGTACCCCTTGTATTTGGGAAGGAATCTGTGC |
| IRF3-luc-R | CCGCTCGAGGGACGGGACCACCCCGA | |
| IFN-β | IFN-β-luc-F | GGGGTACCGCATCCTCCAACACCTCTTCAACAT |
| IFN-β-luc-R | CCGCTCGAGTGGTTTACGAAGCATTGCTCAAGGT |
For luciferase reporter gene assays, promoter plasmids with 1.5-kb DNA fragments encompassing promoter sequences of cGAS, TBK1, STING, IRF7, or IFN-β retrieved from the Ensembl Regulatory Build database were respectively constructed into a pGL3-BASIC vector by standard molecular biology techniques. Oligonucleotide primers used in promoter constructs are listed in Table 1.
Quantitative Real-Time PCR
Total RNA from MSB1 cells was extracted using a commercial kit from TOYOBO Life Science (Shanghai, China). The mRNA levels of relative genes were performed using real-time quantitative PCR (qPCR) master mix (DBI Bioscience, Germany) on the ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA) using the primers in Table 2. Data shown are the relative abundance of the indicated mRNA normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in each sample. All data were analyzed using the comparative cycle threshold (2−ΔΔCT) method (Livak and Schmittgen, 2001).
Table 2.
Oligonucleotide sequences for RT-qPCR.
| Gene | Name | Oligonucleotide sequences (5′-3′) |
|---|---|---|
| CAV-VP1 | qVP1-F | CTCAAGCGACTTCGACGAAG |
| qVP1-R | AGCCTCACACTATACGTACCG | |
| IFN-β | qIFN-b-F | GCATCCTCCAACACCTCTTCAACAT |
| qIFN-b-R | TGGTTTACGAAGCATTGCTCAAGGT | |
| STING | qSTING-F | ATCCAGTACCTGGCAGACCT |
| qSTING-R | ACAAGAAGTGGCTCTCAGGC | |
| cGAS | qcGAS-F | GAGATGGACAACCGCTACG |
| qcGAS-R | TCTGCACCACGTACCTGTCC | |
| GAPDH | qGAPDH-F | CGATCTGAACTACATGGTTTAC |
| qGAPDH-R | TCTGCCCATTTGATGTTGC |
Cell transfection and Luciferase Reporter Assays
Interferon stimulatory DNA (ISD), a 45-bp non-CpG oligomer, can strongly enhance the expression of IFN-β in various cell types (Stetson and Medzhitov, 2006; Ishikawa et al., 2009). ISD (InvivoGen, San Diego, CA) was applied as a simulator of IFN-β production for determining the effect of VP1 on IFN-β. Briefly, 5 × 104 cells were seeded onto 24-well plates and transiently transfected with the firefly luciferase reporter plasmid (100 ng), pRL-TK-Renilla luciferase reporter (50 ng), and indicated plasmids or pCMV-myc empty control vector (500 ng) using Lipofectamine 3000 from Thermo Fisher Scientific (Shanghai, China), with the STING plasmid or 2.5 μg of ISD transfection as a simulator.
After 24 h, cells were lysed, and samples were assayed with the dual luciferase reporter assay kit (Vazyme, Nanjing, China) and measured by a multitube chemiluminescence detector (FilterMax F3/F5, Molecular Devices). The reporter assays were repeated at least 3 times.
SDS-PAGE, Native PAGE, and Western Blot Analysis
The expression plasmids were transfected into the indicated cells. Thirty-six hours post-transfection, cells were lysed on ice with 300 µL of prechilled RIPA buffer (Servicebio, Wuhan, China) containing 1 mM PMSF (Solarbio Science & Technology, Beijing, China). The lysate was centrifuged for 10 min at 12,000 × rpm and subjected to SDS-PAGE or native PAGE. For western blot analysis, samples were separated by electrophoresis on SDS-PAGE or native PAGE, transferred to polyvinylidene difluoride (PVDF) membranes and incubated with the indicated primary antibodies and peroxide-conjugated secondary antibodies. A chemiluminescence system was used to detect proteins according to the manufacturer's protocol.
Co-Immunoprecipitation
Cells were co-transfected with the indicated expression plasmids. The sample was extracted and incubated with 25 µL anti-Myc affinity magnetic beads (Bio-Linkedin, Shanghai, China) overnight at 4°C. The beads were washed 3 times with Tris-buffered saline with 0.05% Tween 20 (TBST) and boiled with 5 × SDS loading buffer for 8 min before analysis by western blotting with the indicated antibodies.
Confocal Microscopy and Subcellular co-Localization
Chicken DF-1 cells were co-transfected with the expression plasmid pmCherry-VP1 (1.0 µg) together with pEGFP-IRF7 (1.0 µg) or control plasmids (1.0 µg). For confocal imaging, the cells were fixed with 4% paraformaldehyde for 20 min and then counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Solarbio Science & Technology, Beijing, China) to visualize cell nuclei. After washing PBS 2 times, the cells were examined using a confocal microscope (CKX41-32FL, Olympus, Japan).
Statistical Analysis
Data were expressed as means ± standard deviations. Statistical significance between groups was determined using Student's t-test or one-way ANOVA as mentioned in the figure legends with GraphPad Prism 8.0 software. P-values are indicated in figures with asterisks as *P < 0.05, **P < 0.01, ***P < 0.001, and ns indicates no significant difference.
RESULTS
CAV Infection Activates cGAS and STING but Inhibits IFN-β Expression
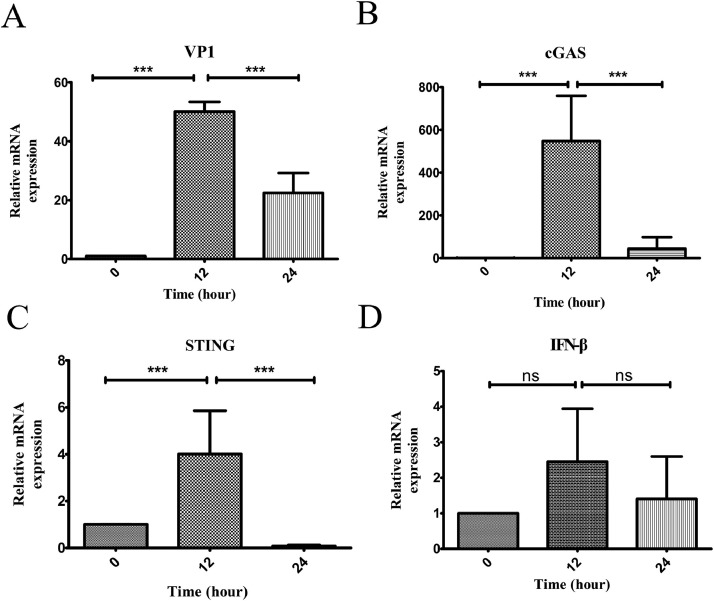
To determine whether CAV infection affects cGAS, STING, and type I IFN gene expression, we used real-time qPCR to analyze virus infection-induced mRNA levels of cGAS, STING, and IFN-β in MSB1 cells at 0, 12, and 24 h postinfection (hpi). The viral capsid VP1 gene was used as a viral replication indicator (Figure 1A). With virus infection, mRNA expressions of cGAS (Figure 1B) and STING (Figure 1C) significantly increased and peaked at 12 hpi and then declined. In contrast, IFN-β expression level was elevated after CAV infection, but not significantly (Figure 1D). These results indicate that CAV infection could stimulate cGAS and STING but inhibits IFN-β expression.
Figure 1.
CAV infection activates cGAS-STING but inhibits IFN-β expression in cells. Levels of cGAS, STING, and IFN-β mRNA in MSB1 cells were measured by real-time qPCR at the indicated times post-CAV infection, and their mRNA levels were normalized to the GAPDH mRNA level in each sample. Data are presented as means ± standard deviations from at least three independent experiments; ***P < 0.001, ns: no statistical significance.
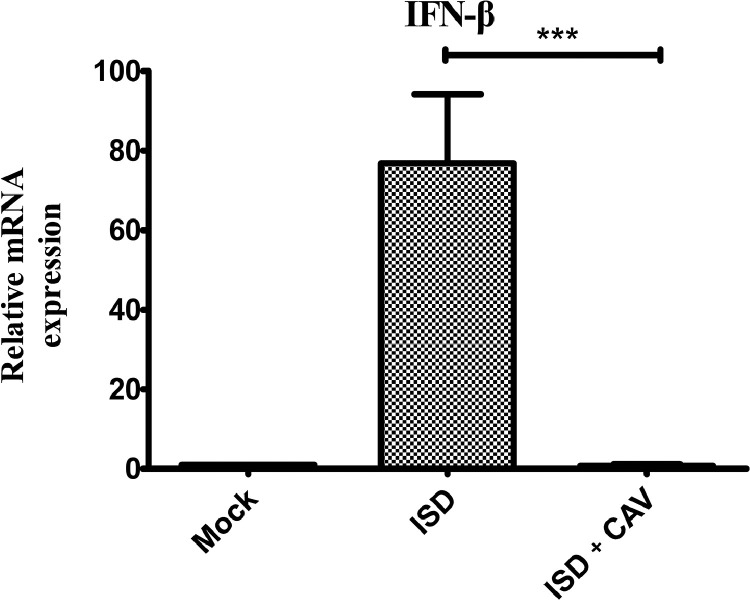
We next considered whether the STING pathway is relevant to IFN-β expression after CAV infection. We stimulated MSB1 cells with ISD, a non-CpG DNA oligomer that activates the STING pathway (Ishikawa et al., 2009), following the CAV-infected cells and mock-infected cells. As shown in Figure 2, we found that the IFN-β mRNA level was strongly promoted in virus mock-infected cells, but such promotion disappeared with CAV infection, suggesting that the virus infection inhibited IFN-β expression via signal transfer of the cGAS-STING signaling axis.
Figure 2.
CAV infection blocks ISD-mediated IFN-β expression. Following incubation with 1 × 104 TCID50 CAV for 3 h, cells were transfected with or without ISD before RT-qPCR. At 6 h post ISD transfection, cells were harvested at 6 h and evaluated for the level of IFN-β mRNA using RT-qPCR. Data are presented as mean ± standard deviations from at least three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
Viral VP1 Disrupts IFN-β Activation by Inhibiting the cGAS-STING Pathway
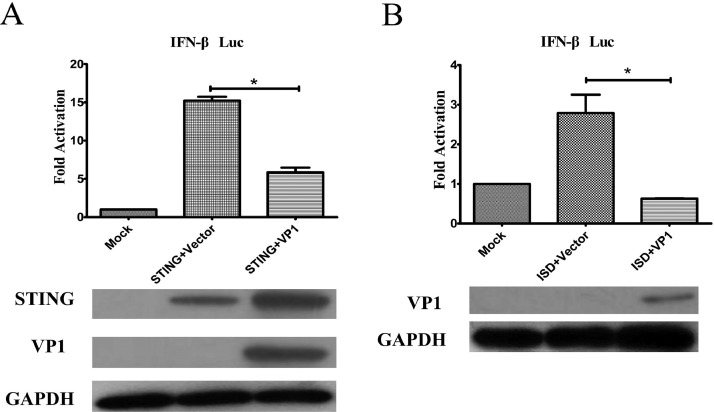
VP1 is the sole structural protein of CAV. We next determined whether VP1 affects IFN-β activation. In this study, STING or ISD was used as a stimulator to determine whether VP1 affects IFN-β production. As shown in Figure 3, reporter assays indicated that VP1 expression could significantly hamper STING or ISD-induced activation of the IFN-β promoter (Figure 3A and B). These results further suggest that VP1 impairs the downstream signal transduction of cGAS-STING pathway mediated IFN-β expression.
Figure 3.
Identification of VP1 as an Inhibitor of STING or ISD-stimulated activity of the IFN-β promoter. (A) HD11 cells were transfected with IFN-β-Luc reporter and STING expression plasmid, along with a pCMV-Myc control plasmid or VP1 construct. (B) Cells were transfected with IFN-β-Luc reporter and the stimulator ISD, together with a pcDNA3.1 empty vector or VP1 construct. Cells were harvested at 24 h after transfection and IFN-β luciferase activity was analyzed. All data represent results from one of three independent experiments and are plotted as means ± standard deviations from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
VP1 Impairs IFN-β Activation via Targeting IRF7
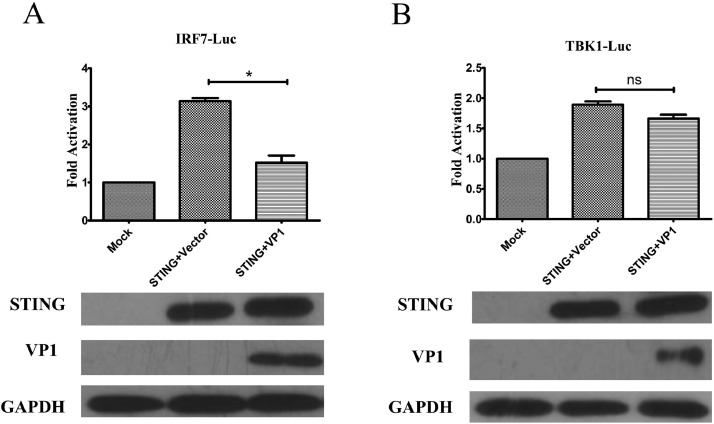
To examine the effect of VP1 on the cGAS-STING signal axis, we next tested whether VP1 directly interacts with downstream adaptors, IRF7, and TBK1 of the cGAS-STING pathway. DF-1 fibroblast cells were co-transfected with TBK1 or IRF7 reporter plasmids together with expression plasmids for STING and VP1 or the control vector. Consistent with previous results, overexpression of VP1 inhibited STING-induced IRF7 activation (Figure 4A), whereas there was no inhibition observed with STING-induced TBK1 activation (Figure 4B). These results suggested that VP1 could impair IFN-β expression via targeting the IRF7 of the cGAS-STING pathway signal axis.
Figure 4.
VP1 inhibits IFN-β activation by selectively targeting IRF7. The cells were cotransfected with a pRL-TK-Luc control plasmid, STING expression plasmid with the empty vector or VP1 construct, together with the IRF7-Luc reporter (A) or TBK1-Luc reporter plasmid (B) for 24 h before luciferase assays. All data are presented as the means ± standard deviations from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001, ns indicates comparison.
VP1 Interacts With IRF7 and Disrupts Dimerization
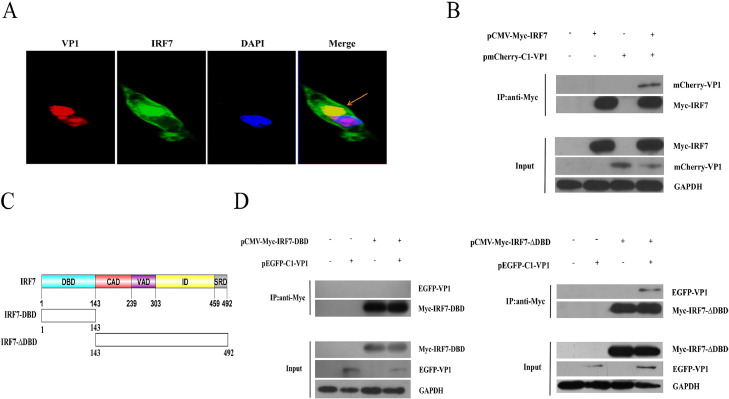
To investigate the molecular mechanisms of the regulatory role of VP1 on IRF7 inhibition, co-localization of VP1 with IRF7 was determined by transient transfection. It has been reported previously that nuclear protein VP1 can partially display in the cytoplasm (Lai et al., 2018). We found that VP1 was co-localized with IRF7 in the cytoplasm (Figure 5A). Coimmunoprecipitation experiments indicated that VP1 interacted with IRF7 (Figure 5B).
Figure 5.
VP1 interacts with IRF7. DF-1 cells were cotransfected with pmCherry-VP1 and pEGFP-IRF7 plasmids. The co-localization of VP1 (red) and IRF7 (green) was detected by confocal microscopy (A), and interactions between VP1 (red) and IRF7 (green) were analyzed by immunoprecipitation with indicated antibodies (B). Schematic representation of the full-length IRF7 (aa 1–492) and truncated IRF7 including IRF7-DBD (aa 1–143) and IRF7-∆DBD (aa 143–492) (C). The cells were transfected with a VP1 plasmid together with the IRF7-DBD or IRF7-∆DBD construct. (D) The cells were cotransfected with VP1 expression plasmid and the indicated IRF7-∆DBD constructs before immunoprecipitation.
Furthermore, we were interested in the region of IRF7 that binds to VP1. There are a DNA-binding domain (DBD, aa 1–43) and a deletion of the DBD (∆DBD, aa 143–492) in IRF7. Previously, it has been shown that C-terminal IRF7-∆DBD retains effector function as well as intact IRF7 in terms of constitutive activation, virus-activated, inhibitory, and signal response (Figure 5C) (Gao et al., 2019). Therefore, we examined whether IRF7-∆DBD binds to VP1. By co-expression of VP1 together with IRF7-DBD or IRF7-∆DBD, coimmunoprecipitation and western blot experiments showed VP1 immunoprecipitated with IRF7-∆DBD, but not with IRF7-DBD (Figure 5D), suggesting that IRF7-∆DBD was essential for the interaction between IRF7 and VP1.
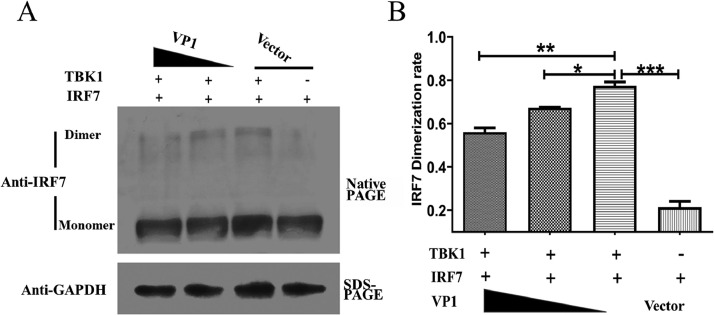
The IRF dimeric form translates to the nucleus where it activates transcription (Wu et al., 2009). To investigate the effect of VP1 on the activity of IRF, we determined the effects of VP1 expression on the dimerization of IRF7. As shown in Figure 6A, the coimmunoprecipitation experiments indicated that overexpression of VP1 clearly attenuated TBK1-induced dimerization of IRF7 in a dose-dependent way (Figure 6B), suggesting that VP1 impairs the activity of IRF7.
Figure 6.
Identification of VP1 as an inhibitor of IRF7 dimerization in a dose-dependent manner. HEK293T cells were transfected with expression plasmids for 2 μg IRF7 together with VP1 in an increased dose from 0, 1, and 2 μg. Twenty-four hours later, cells were incubated with 2 μg TBK1 expression plasmid or the empty vector as a stimulator for another 12 h. The samples were separated by electrophoresis on 12% native PAGE gels, and then analyzed by western blotting (A), the relative dimerization level versus the total level of IRF7 protein was indicated as the average of three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
Previous studies have demonstrated that the cGAS-STING pathways regulate innate immune responses via inhibiting IFN production in response to some dsDNA viruses in chickens (Gao et al., 2019; Wang et al., 2020). Here, we demonstrate for the first time that CAV infection can stimulate cGAS/STING but suppress IFN-β expression in host cells. We further investigated whether CAV proteins target the cGAS-STING axis for immune evasion. Our results showed that the capsid VP1 is recruited to hamper the activation of IFN-β by using ISD or STING as the DNA stimulator. These results suggest that CAV VP1 acts as a negative regulator to interfere with IFN-β production by blocking the cGAS-STING pathway.
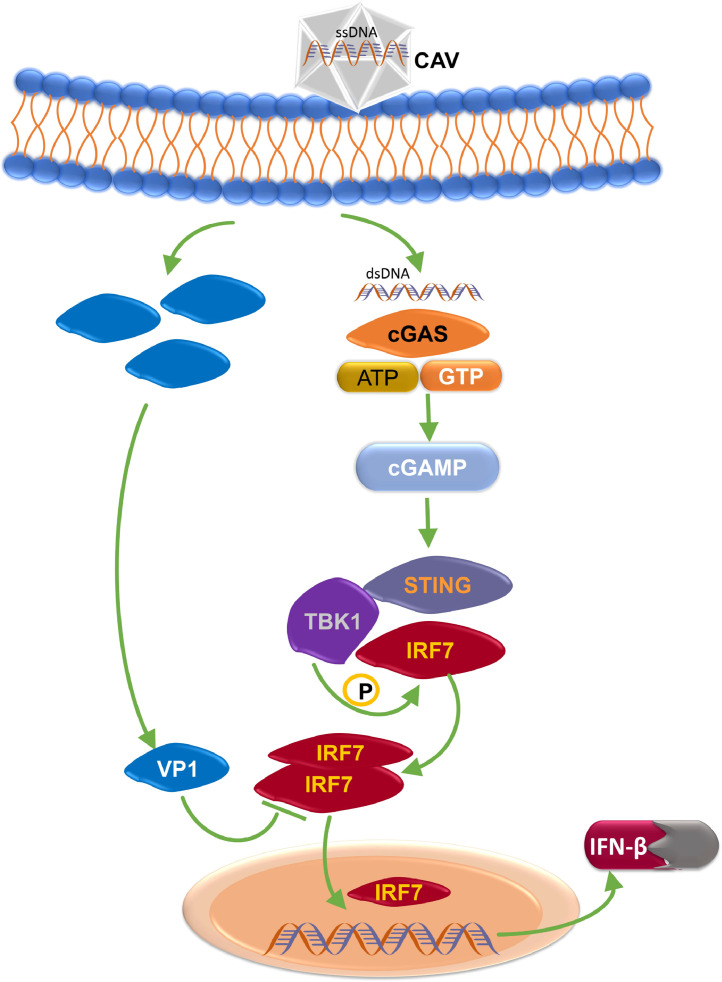
The host organism regulates IRF3 to promote its participation in the formation of the IFN-β enhanceosome (Lin et al., 2000; Schwanke et al., 2020). Therefore, viruses have developed strategies to counteract the innate immune response by targeting IRF3 (Zhang et al. ,2016). For example, HSV-1 encodes both UL46 and ICP27 to hamper IFN production via inhibiting IRF3 activation (Christensen et al. 2016; Deschamps and Kalamvoki, 2017). As chickens lack classical IRF3, IRF7 is thought to participate in STING-mediated IFN-β signaling (Santhakumar et al., 2017; Neerukonda and Katneni, 2020). Here, we identified VP1 as an inhibitor that hampers IFN-β production after CAV infection. Subsequently, our results suggest that VP1 does not affect TBK1 activation, but significantly blocks IRF7 activation. However, the interaction of VP1-TBK1-IRF7 needs further investigation. Previous studies have shown that upon docking to adaptor proteins and subsequent phosphorylation, IRF3/7 is required to form a dimer to translocate to the nucleus and be transcriptionally active (Dalskov et al., 2020). Furthermore, we confirmed that VP1 interacted with IRF7 by selectively binding to IRF7 with a DBD deletion (aa 143–492) which was reported to be essential for the dimerization and phosphorylation of IRF7 (Wang et al., 2020). In our study, we found for the first time that CAV VP1 is a potent inhibitor of the crash-STING pathway that binds to IRF7 and disrupts IRF7 dimerization, which ultimately can reduce IFN-β production and weaken the innate antiviral response (Christensen et al., 2016; Dong et al., 2022) (Figure 7).
Figure 7.
Schematic of the inhibitory effect of CAV VP1 on the cGAS-STING pathway. CAV encoded VP1 can inhibit the production of IFN-β through suppression of IRF7 activation of the cGAS-STING pathway by hampering IRF7 dimerization, resulting in reduced nuclear translocation of IRF7. This leads to the inhibition of IFN-β production, thereby enabling the immune escape of the virus in the host cell.
In summary, our results provide evidence that the chicken cGAS-STING signal pathway plays an important role in regulating IFN-β response to CAV infection. In this study, the CAV VP1 was identified as an effective inhibitor of the cGAS-STING pathway that mediated activation of downstream antiviral genes by interacting with IRF7 and weakening the innate antiviral response. Our findings would allow a better understanding of the interplay between CAV and the host, CAV evasion strategies, and the rationale for vaccine development against CAV infection.
Author contributions
Author contributions: Conceptualization, XL.L. and FF.S.; data curation, XL.L., DX.X, AY. X., Y.W, T.S., TT.M., and H.Y.; funding acquisition, XL.L. and FF.S.; investigation, FZ.X, and H.Z.; supervision, JN.L. and FF.S.; writing–original draft, XL.L. and DX.X.; writing—review and editing, XL.L. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This work was supported by the Anhui Provincial Natural Science Foundation (2108085MC115) and the Natural Science Research Project of Anhui Universities (KJ2018A0136). We thank Dr. Chang Xinyue for helpful suggestions on revising the manuscript.
Disclosures
The authors have no conflict of interest to declare.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102291.
Appendix. Supplementary materials
References
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Ro¨hl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M.H., Jensen S.B., Miettinen J.J., Luecke S., Prabakaran T., Reinert L.S., Mettenleiter T., Chen Z.J., Knipe D.M., Sandri-Goldin R.M., Enquist L.W., Hartmann R., Mogensen T.H., Rice S.A., Nyman T.A., Matikainen S., Paludan S.R. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO. J. 2016;35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalskov L., Narita R., Andersen L.L., Jensen N., Assil S., Kristensen K.H., Mikkelsen J.G., Fujita T., Mogensen T.H., Paludan S.R., Hartmann R. Characterization of distinct molecular interactions responsible for IRF3 and IRF7 phosphorylation and subsequent dimerization. Nucleic Acids Res. 2020;48:11421–11433. doi: 10.1093/nar/gkaa873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps T., Kalamvoki M. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J. Virol. 2017;91 doi: 10.1128/JVI.00535-17. e00535–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.V., Tran G.T.H., Kurokawa A., Yamamoto Y., Takeda Y., Ogawa H., Imai K. Genetic characterization of chicken anemia viruses newly isolated from diseased chicks in Japan in 2020. J. Vet. Med. Sci. 2022;84:166–170. doi: 10.1292/jvms.21-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.Z., Su S., Gao Y.Q., Wang P.P., Huang Z.F., Hu M.M., Luo W.W., Li S., Luo M.H., Wang Y.Y., Shu H.B. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe. 2017;21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Gao L., Li K., Zhang Y., Liu Y., Liu C., Zhang Y., Gao Y., Qi X., Cui H., Wang Y., Wang X. Inhibition of DNA-sensing pathway by Marek's disease virus VP23 protein through suppression of interferon regulatory factor 7 activation. J. Virol. 2019;93 doi: 10.1128/JVI.01934-18. e01934–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giotis E.S., Scott A., Rothwell L., Hu T., Talbot R., Todd D., Burt D.W., Glass E.J., Kaiser P. Chicken anaemia virus evades host immune responses in transformed lymphocytes. J. Gen. Virol. 2018;99:321–327. doi: 10.1099/jgv.0.001011. [DOI] [PubMed] [Google Scholar]
- Huang L., Xu W., Liu H., Xue M., Liu X., Zhang K., Hu L., Li J., Liu X., Xiang Z., Zheng J., Li C., Chen W., Bu Z., Xiong T., Weng C. African Swine Fever Virus pI215L negatively regulates cGAS-STING signaling pathway through recruiting RNF138 to inhibit K63-linked ubiquitination of TBK1. J. Immunol. 2021;207:2754–2769. doi: 10.4049/jimmunol.2100320. [DOI] [PubMed] [Google Scholar]
- Ishii K.J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H., Sato S., Yamamoto M., Uematsu S., Kawai T., Takeuchi O., Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai G.H., Lin M.K., Lien Y.Y., Cheng J.H., Sun F.C., Lee M.S., Chen H.J., Lee M.S. Characterization of the DNA binding activity of structural protein VP1 from chicken anaemia virus. BMC Vet. Res. 2018;14:155. doi: 10.1186/s12917-018-1465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Mamane Y., Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- Liu X.L., Shan W.J., Jia L.J., Yang X., Zhang J.J., Wu Y.R., Xu F.Z., Li J.N. Avian leukosis virus subgroup J triggers caspase-1-mediated inflammatory response in chick livers. Virus Res. 2016;215:65–71. doi: 10.1016/j.virusres.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Motwani M., Mcgowan J., Antonovitch J., Gao M.J., Jiang Z., Sharma S. cGAS-STING pathway does not promote autoimmunity in murine models of SLE. Front. Immunol. 2021;8:689. doi: 10.3389/fimmu.2021.605930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerukonda S.N., Katneni U. Avian pattern recognition receptor sensing and signaling. Vet. Sci. 2020;7:14. doi: 10.3390/vetsci7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhakumar D., Rubbenstroth D., Martinez-Sobrido L., Munir M. Avian interferons and their antiviral effectors. Front. Immunol. 2017;8:49. doi: 10.3389/fimmu.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K.A. Chicken anemia virus. Curr. Top Microbiol. Immunol. 2009;331:151–183. doi: 10.1007/978-3-540-70972-5_10. [DOI] [PubMed] [Google Scholar]
- Schwanke H., Stempel M., Brinkmann M.M. Of keeping and tipping the balance: Host regulation and viral modulation of IRF3-Dependent IFNB1 expression. Viruses. 2020;12:733. doi: 10.3390/v12070733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C., Li X., Li P. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev. 2014;25:641–648. doi: 10.1016/j.cytogfr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M.K., Nandakumar R., Stadler D., Malo A., Valls R.M., Wang F., Reinert L.S., Dagnaes-Hansen F., Hollensen A.K., Mikkelsen J.G., Protzer U., Paludan S.R. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology. 2016;64:746–759. doi: 10.1002/hep.28685. [DOI] [PubMed] [Google Scholar]
- Wang J., Ba G., Han Y.Q., Ming S.L., Wang M.D., Fu P.F., Zhao Q.Q., Zhang S., Wu Y.N., Yang G.Y., Chu B.B. Cyclic GMP-AMP synthase is essential for cytosolic double-stranded DNA and fowl adenovirus serotype 4 triggered innate immune responses in chickens. Int. J. Biol. Macromol. 2020;146:497–507. doi: 10.1016/j.ijbiomac.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Wani M.Y., Dhama K., Malik Y.S. Impact of virus load on immunocytological and histopathological parameters during clinical chicken anemia virus (CAV) infection in poultry. Microb. Pathog. 2006;96:42–51. doi: 10.1016/j.micpath.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., Chen Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fossum E., Joo C.H., Inn K.S., Shin Y.C., Johannsen E., Hutt-Fletcher L.M., Hass J., Jung J.U. Epstein-Barr virus LF2: an antagonist to type I interferon. J. Virol. 2009;83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Gao P., Gao G., Fan Z. DNA sensor cGAS-mediated immune recognition. Protein Cell. 2016;7:777–791. doi: 10.1007/s13238-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Su C., Zheng C. Herpes Simplex Virus 1 Serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J. Virol. 2016;90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.