Figure 3.
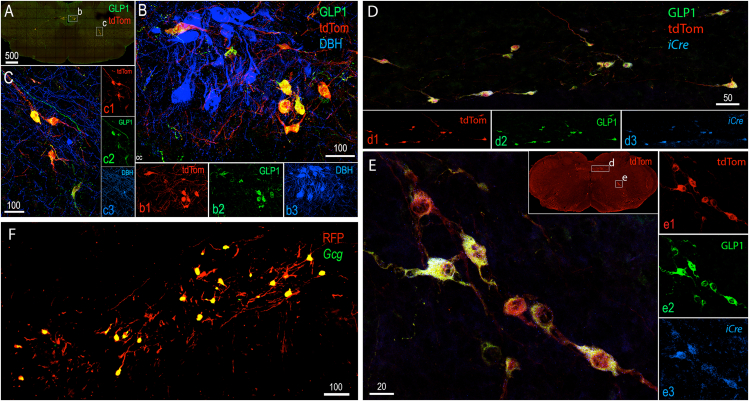
Efficiency and selectivity of hindbrain iCre expression in Gcg-Cre/tdTom reporter rats. A, low magnification view of the hindbrain distribution of tdTom (red) and GLP1 (green) immunolabeling. The boxed regions (b, c) are shown in higher-magnification confocal images in panels B and C. B, Blue (DBH-positive) noradrenergic neurons in the cNTS are not tdTom-positive. All tdTom-positive cNTS neurons are GLP1-positive, and vice-versa. C, Blue (DBH-positive) noradrenergic neurons in the IRt are not tdTom-positive. All tdTom-positive IRt neurons are GLP1-positive, and vice-versa. D, RNAscope FISH reveals iCre (blue) mRNA expression in tdTom/GLP1 positive neurons within the cNTS (D) and within the IRt (panel E). Boxed regions shown in the inset in panel E are depicted in higher-magnification confocal images in panels D and E. F, saggital section through the cNTS depicting overlap of RFP immunolabeling (labeling tdTom + cells and processes) and Gcg mRNA expression (green, RNAscope). All RFP-positive cell bodies within the cNTS express Gcg. Scale bars are in microns.