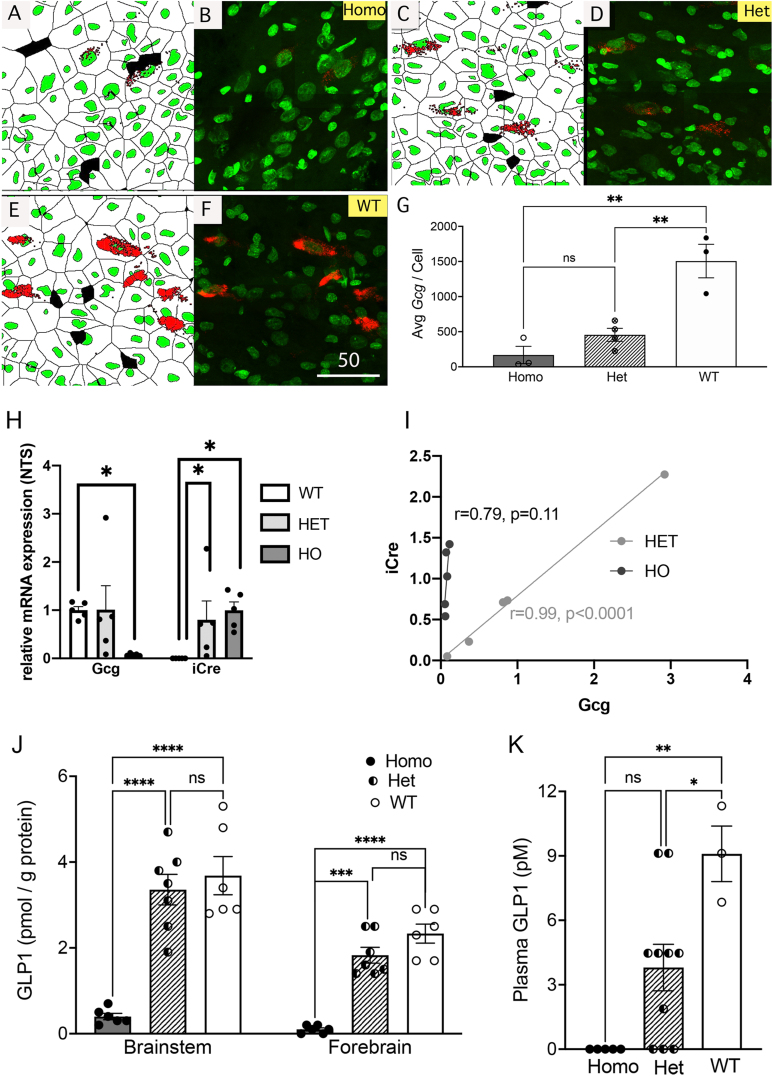
Figure 5.
Quantification of Gcg mRNA expression and GLP1 protein levels in Gcg-Cre rats. A-F, RNAscope FISH followed by HALO-based image analysis of cellular Gcg mRNA transcript levels within the cNTS. A + B, Homo Gcg-Cre rats display very low cellular levels of Gcg mRNA (red). C + D,Gcg mRNA transcript labeling (red) in Het Gcg-Cre rats. E+F,Gcg mRNA transcript labeling (red) in WT Gcg-Cre rats. G, summary data depicting the average number of Gcg mRNA transcripts quantified in Homo, Het, and WT Gcg-Cre rats. Cellular levels of Gcg mRNA in Homo and Het rats are lower than in WT rats (∗∗P < 0.01 for each comparison). H, qRT-PCR analysis of Gcg and iCre mRNA expression within the NTS. Gcg mRNA expression in Het rats (light gray bars) is not significantly different compared to WT rats, whereas Gcg mRNA expression in Homo rats (Ho, dark gray) is nearly undetectable. WT rats lack detectable iCre expression, whereas iCre mRNA levels are similar in Het and Homo rats. I, within-subjects correlation of iCre and Gcg mRNA expression within the NTS in Het and Homo rats. Expression levels are variable but tightly correlated in Het rats, but are not correlated in Homo rats, which have barely detectable levels of Gcg mRNA expression. J, GLP1 protein levels analyzed via ELISA in tissue samples from the brainstem and forebrain in Gcg-Cre rats. Brainstem and forebrain GLP1 protein levels are similar in Het and WT Gcg-Cre rats, but are markedly lower in Homo Gcg-Cre rats (∗∗P < 0.001). K, Plasma levels of GLP1 protein analyzed via ELISA in Gcg-Cre rats. Compared to plasma GLP1 levels in WT rats, levels in Het rats are reduced by approximately 50%, and levels in Homo rats are essentially undetectable (∗P < 0.05; ∗∗P < 0.01).