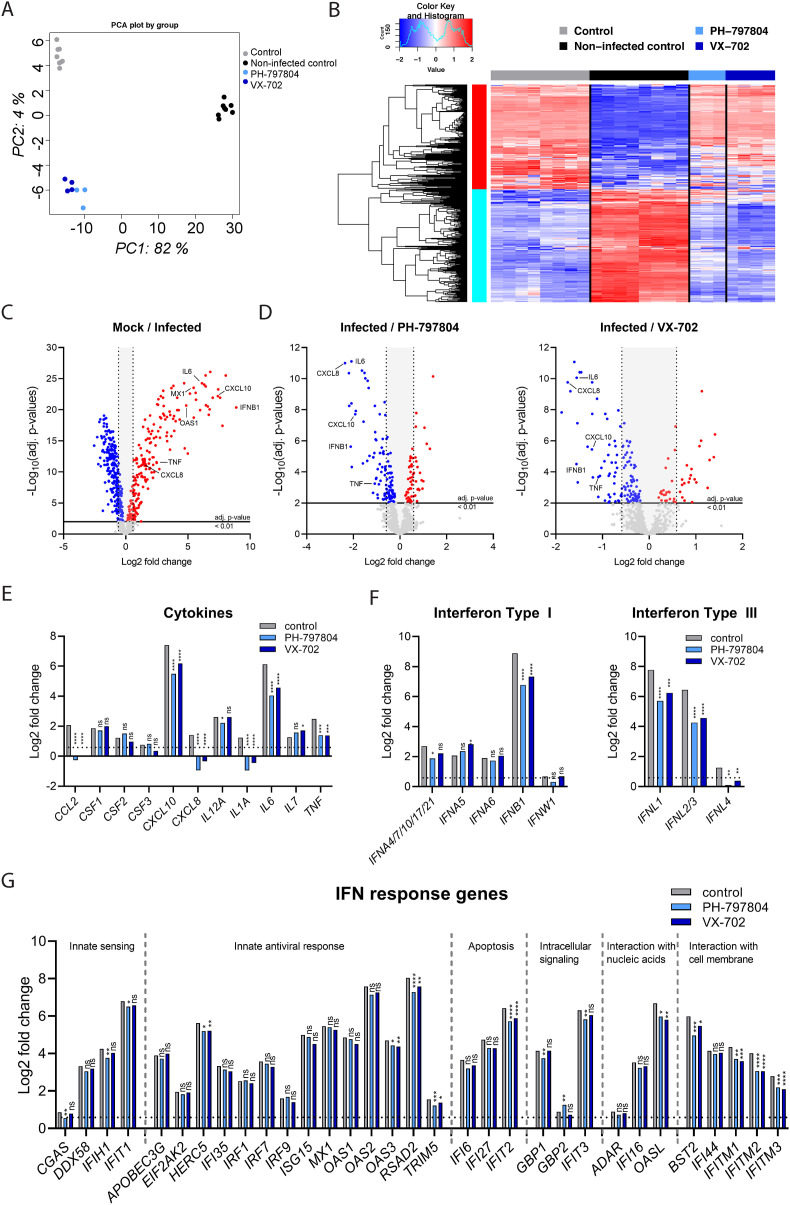
Fig. 3.
p38 MAPK inhibitors PH-797804 and VX-702 reduce the inflammatory response to SARS-CoV-2. Calu-3 cells were treated for 1 h with 5 μM of PH-797804, 5 μM of VX-702 or DMSO (control) before infection with SARS-CoV-2 at MOI 0.001 for 48 h in the presence of the inhibitors. Non-infected, DMSO-treated cells were used as an additional control (non-infected control, mock). Gene expression was analyzed using the multiplexed RNA hybridization NanoString Human Host Response Panel and compared between non-treated and infected cells as well as infected and inhibitor-treated cells to determine differentially expressed genes (DEG). Statistical significance was determined using multiple testing and Benjamini and Hochberg correction. A) Principal component analysis (PCA) of relative gene expression levels from all samples. B) Heatmap representation of gene expression levels in mock (n = 6), solvent control (n = 6) and SARS-CoV-2 infected samples treated with PH-797804 (n = 3) and VX-702 (n = 4). C) Volcano plot of up- and down-regulated host response genes during SARS-CoV-2 infection; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). D) Effect of inhibitor treatment on host response genes compared to infected cells using linear regression from the package LIMMA; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). Comparison of mRNA expression in infected and inhibitor-treated infected cells of E) COVID-19-relevant pro-inflammatory cytokines, F) Type I, II and III interferons, G) IFN response genes normalized to the non-infected control. Data are expressed as means of Log2-fold changes; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). *p < 0.05; **p < 0.01, ***p < 0.001.