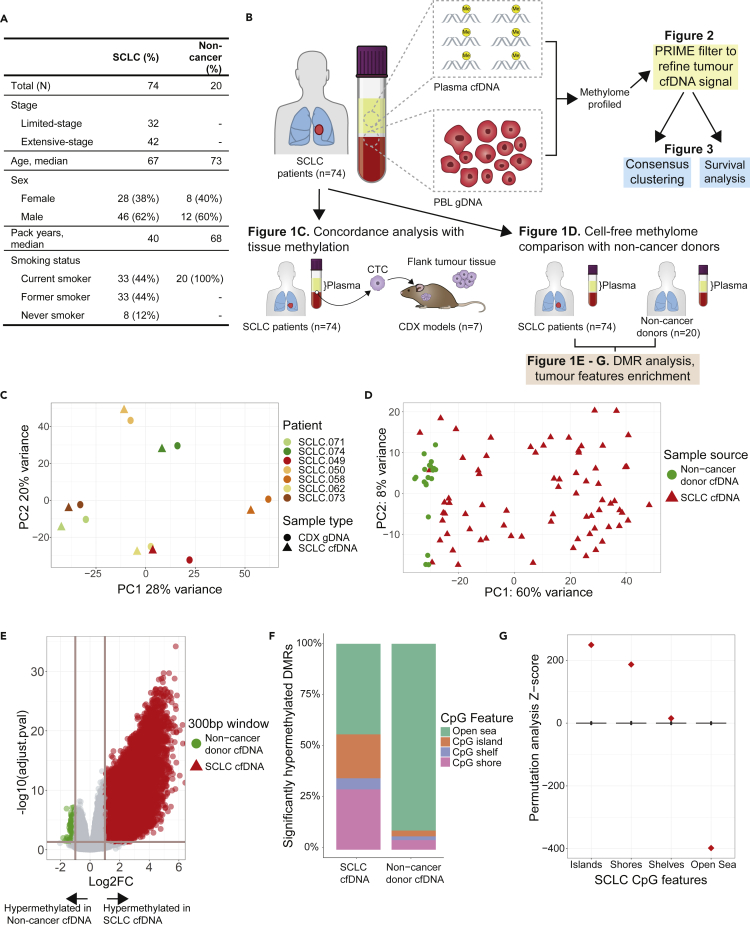
Figure 1.
SCLC cell-free methylome reflects tissue and is distinct from non-cancer
(A) Demographics and clinical characteristics of 74 patients with small-cell lung cancer (SCLC) and non-cancer donors.
(B) Schematic of overall structure of study and major analyses done.
(C) Principal component analysis of genome-wide methylation profiles of total methylated cell-free DNA (cfDNA) from patients with SCLC (n = 7) and methylated genomic DNA from paired circulating tumor cell-derived xenograft (CDX) models (n = 7).
(D) Principal component analysis of genome-wide methylation profiles of total methylation cfDNA from patients with SCLC (n = 74) and non-cancer donors (n = 20).
(E) Volcano plot of differentially methylated region (DMR) analysis of patients with SCLC (n = 74) and non-cancer donors (n = 20). Each dot corresponds to a 300bp region of the genome. The horizontal line corresponds to p-adjusted = 0.05 and vertical lines correspond to log2 fold-change of +/−2. There are 51,666 hypermethylated DMRs in SCLC and 1,019 in non-cancer donors.
(F) Bar plot of significantly hypermethylated DMRs corresponding to CpG features observed in patients with SCLC (n = 51,666 DMRs) and non-cancer donors (n = 1,019 DMRs).
(G) Permutation analysis of the hypermethylated DMRs observed in SCLC (n = 51,666 DMRs).