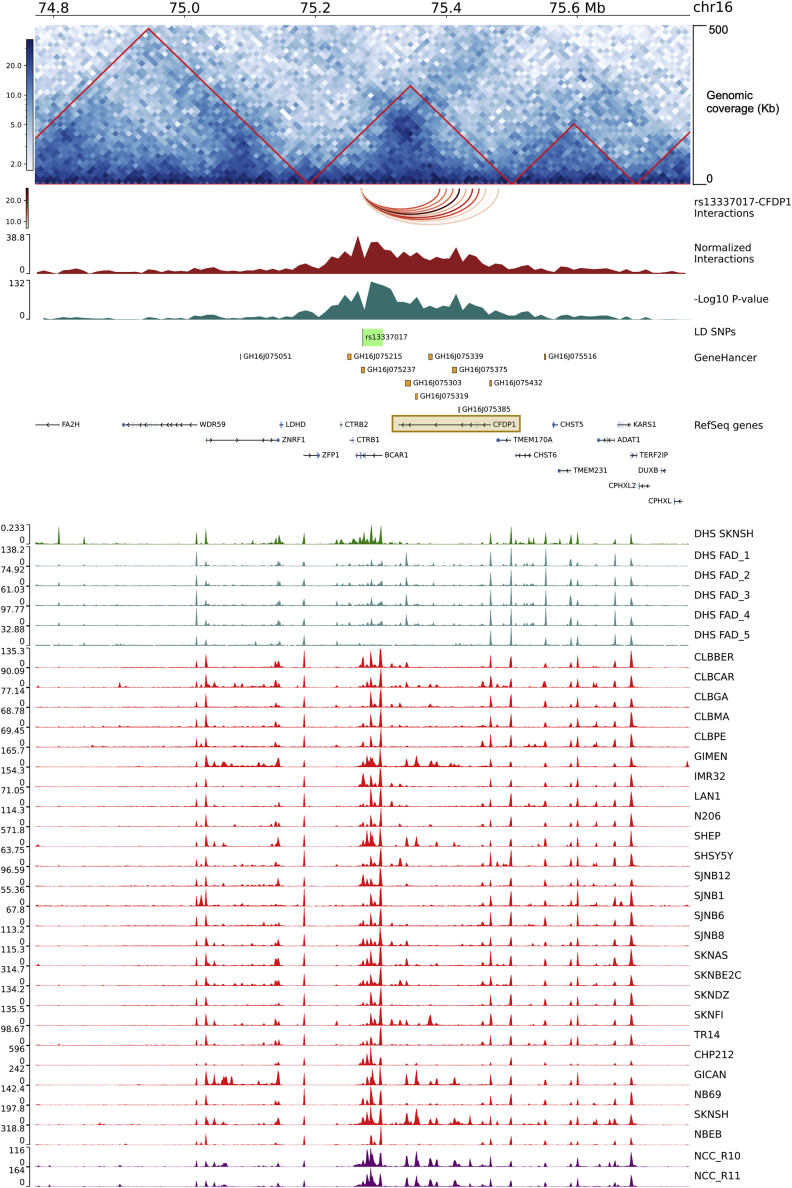
Figure 2.
rs13337017 showed long-range significant interactions with CFPD1 in SK-N-BE NB cell line
The figure reports the genomic interactions from the rs13337017 point of view. The genomic tracks are named from top to bottom and described below. The genomic coordinates are from human genome hg19. The interaction matrix is centered on rs13337017 (chr16:75,272,366-75272367) and extended of 0.5 Mb up- and downstream. Genomic coverage is of 500 Kb and the matrix resolution is of 10 Kb (the interactions are calculated between bins of 10 Kb). Red-bordered triangles represent the Topologically Associated Domains (TADs). The arcs track shows the interactions between rs13337017 and CFPD1 annotated bins. The normalized number of interactions (a measure of the strength of interactions). The minus Log10 of the FDR adjusted p value. The region of 30,631 bp containing the list of 58 in Linkage Disequilibrium SNPs (LD SNPs) with rs13337017 is in green. The GeneHancer track reports the regulatory elements, from the GeneHancer database, showing strong associations with CFDP1 (see materials and methods). The NCBI RefSeq genes (we plotted the longest transcript for each gene). A brown-bordered rectangle highlights the CFDP1 locus. The ChIP peak-based tracks show, from top to bottom, the DNase-I hypersensitivity levels (DHS) sites of the SK-N-SH NB cell line in dark green (from ENCODE v3); the DHS signal of fetal adrenal gland (n = 5) tissues from Roadmap Epigenomics Consortium (GEO: GSM530653, GSM817165, GSM1027310, GSM1027311, GSM817167) in dark cyan; the H3K27ac data of NB cell lines (from GEO: GSE90683 and GSE65664) in red; and H3K27ac peaks from two human neural crest cells (NCCs) (from GEO: GSE90683) in purple. Data ranges and heatmap color keys are shown on the left of each track, when needed.