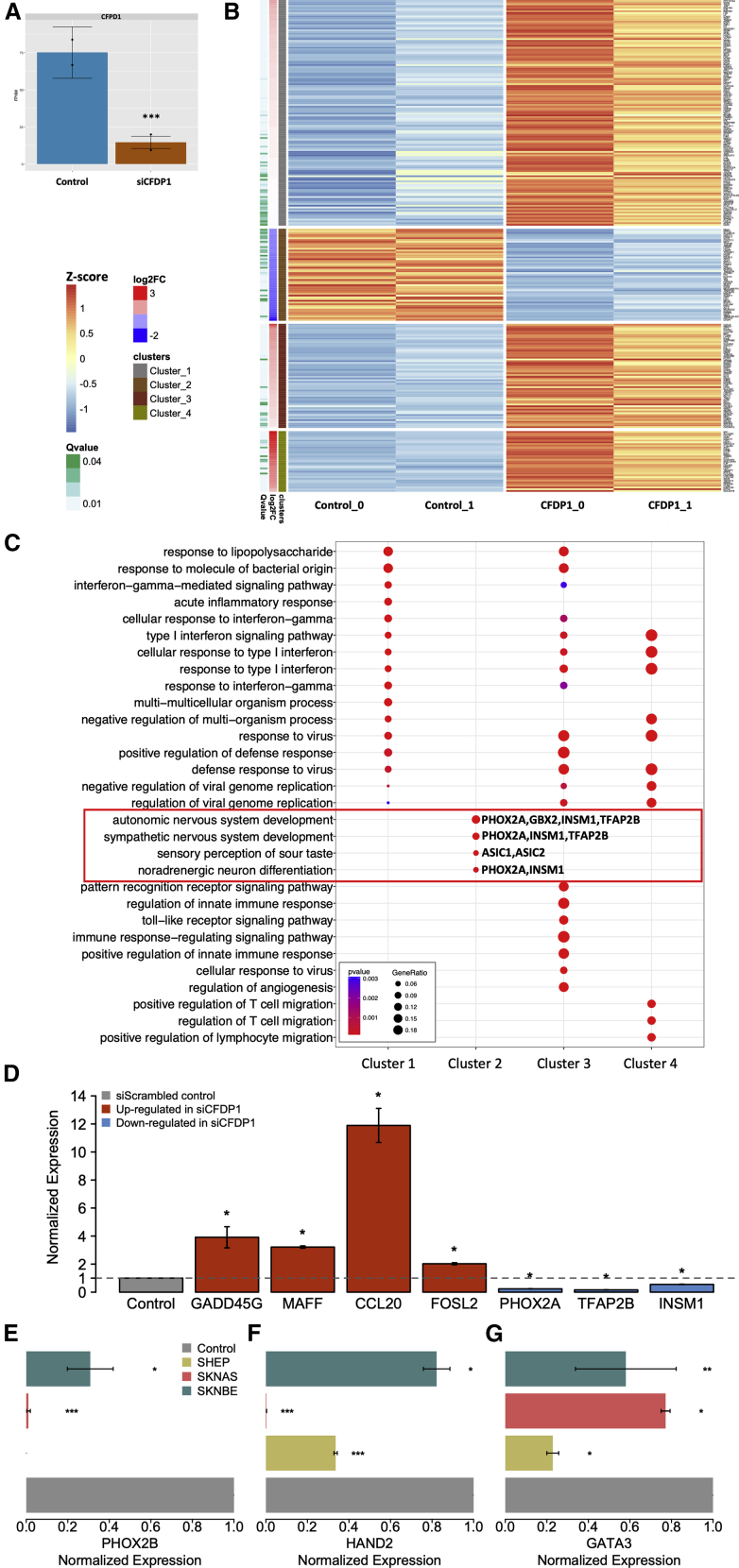
Figure 5.
The depletion of CFDP1 affects the expression of gene regulators of NB cell identity
(A) Barplots showing the expression levels of CFDP1 in control (siScrambled) and CFDP1 silenced cells (siCFDP1) as measured by RNA-seq in SK-N-AS.
(B) Heatmap of the differentially expressed genes grouped according to the k-means clustering algorithm. Color key on the left.
(C) Dotplots reporting the Gene Ontology biological process enrichment results of clustered gene lists.
(D) Barplot showing the results of expression quantification by quantitative real-time PCR for four up-regulated genes (red) and three down-regulated genes (blue) in SK-N-AS.
(E) Expression quantification by quantitative real-time PCR of PHOX2B in SK-N-AS and SK-N-BE NB cell lines.
(F) Expression quantification by quantitative real-time PCR of HAND2 in SK-N-AS, SK-N-BE, and SH-EP NB cell lines.
(G) Expression quantification by quantitative real-time PCR of GATA3 in SK-N-AS, SK-N-BE, and SH-EP NB cell lines. The data in (D) to (G) are reported as fold-changes of induction in siCFDP1 compared with control cells. In Panel (A), (D), (E), (F), and (G) statistical significance was assessed using a t test. ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001.