Abstract
Upregulation of functional network connectivity in the presence of structural degeneration is seen in the premanifest stages of Huntington’s disease (preHD) 10–15 years from clinical diagnosis. However, whether widespread network connectivity changes are seen in gene carriers much further from onset has yet to be explored.
We characterized functional network connectivity throughout the brain and related it to a measure of disease pathology burden (CSF neurofilament light, NfL) and measures of structural connectivity in asymptomatic gene carriers, on average 24 years from onset. We related these measurements to estimates of cortical and subcortical gene expression.
We found no overall differences in functional (or structural) connectivity anywhere in the brain comparing control and preHD participants. However, increased functional connectivity, particularly between posterior cortical areas, correlated with increasing CSF NfL level in preHD participants. Using the Allen Human Brain Atlas and expression-weighted cell-type enrichment analysis, we demonstrated that this functional connectivity upregulation occurred in cortical regions associated with regional expression of genes specific to neuronal cells. This relationship was validated using single-nucleus RNAseq data from post-mortem Huntington’s disease and control brains showing enrichment of neuronal-specific genes that are differentially expressed in Huntington’s disease.
Functional brain networks in asymptomatic preHD gene carriers very far from disease onset show evidence of upregulated connectivity correlating with increased disease burden. These changes occur among brain areas that show regional expression of genes specific to neuronal GABAergic and glutamatergic cells.
Keywords: Huntington’s disease, connectivity, gene expression, neurofilament light
McColgan et al. report that premanifest Huntington’s disease gene carriers around 20 years from disease onset display upregulated functional connectivity that correlates with increasing CSF neurofilament light. This occurs in brain regions that show gene expression specific to neuronal GABAergic and glutamatergic cells.
Introduction
The earliest, asymptomatic stages of neurodegenerative disease involve a complex interplay between the effects of pathology on brain structure and function. Huntington’s disease is a monogenic, autosomal-dominant neurodegenerative disorder where both grey and white matter structural brain changes occur many years prior to disease onset.1–4 Alongside this, functional brain changes also occur,5–11 which may reflect the presence of pathological changes in connectivity12 or compensation in the form of upregulated brain network activity.13–15 However, the biological basis of these changes is unclear. Here, we investigate Huntington’s disease-related functional network connectivity during very early premanifest Huntington’s disease (preHD) in the context of largely intact structural white matter networks using regional gene expression and post-mortem Huntington’s disease single-nucleus RNA sequencing (snRNAseq) data.
Network connectivity in preHD gene carriers is upregulated within functional networks where structural connectivity is weakest.12 As such, marked axonal loss may lead to upregulated functional network connections unaffected by structural change or the recruitment of extra-network functional connections in those around 10–15 years from disease onset.1,16–18 We have previously shown that there are no detectable changes in structural network connectivity in a young-adult preHD cohort (HD-YAS) on average 24 years from disease onset,19,20 but there was evidence of functional network change.11 This is consistent with considerable evidence of neuronal network hyperexcitability driven by glutamatergic excitotoxicity and/or reduced inhibitory GABAergic activity in the earliest stages of neurodegeneration.21–25 However, our earlier study focused specifically on fronto-striatal circuits associated with cognitive flexibility and it is not known whether such functional network changes are more widespread. Determining this is important, because the period more than 20 years from clinical diagnosis is a point at which therapeutic treatments could potentially stall or eliminate disease progression.
Gene-expression profiles underlying patterns of functional connectivity have been investigated in healthy controls,26 neuropsychiatric cohorts27,28 and recently in Parkinson’s disease, where differential patterns of gene expression are associated with decoupling of structural and functional networks.29 In Huntington’s disease, gene transcription levels for synaptic signalling (particularly in the caudate and motor cortex) and cellular metabolism are atypical in human and animal models.30,31 Regions that show degeneration of white matter connections in preHD, around 15 years from disease onset, are also those that exhibit regional expression of synaptic and metabolic genes, particularly those that show abnormal transcription in post-mortem human and animal Huntington’s disease models.18 It is unclear, however, if brain network changes very far from disease onset show the same biological relationships and thus share a common pathobiology with later stage preHD or whether they are driven by different biological mechanisms.
In the current study, we characterized functional network connectivity in a cohort of asymptomatic young adult preHD gene carriers on average 24 years from disease onset.19 First, we sought to characterize pathology-related structural and functional network connectivity. We employed network-based statistics (NBS) to explore differences in network connectivity between preHD and controls and then tested the extent to which any changes were associated with Huntington’s disease pathology in terms of elevated CSF neurofilament light (NfL) levels, a marker of axonal degeneration that correlates with Huntington’s disease progression.32–34 We then investigated the possible mechanisms of any changes in connectivity using Allen Human Brain Atlas (AHBA) regional gene expression data and partial least squares regression. This provided ranked gene lists associated with regions that showed increased functional connectivity. We used these to perform gene ontology (GO) enrichment analyses to identify biological relationships and expression-weighted cell-type enrichment (EWCE) analyses to identify cell-specific relationships. Finally, the regional relationships we observed were validated using both differential gene-expression data from Huntington’s disease animal models and post-mortem Huntington’s disease brains and cell-specific snRNAseq data from Huntington’s disease and healthy post-mortem brains.
Materials and methods
Participants
Sixty-four preHD and 67 control participants matched for age, sex and education were recruited for the Huntington’s disease–Young Adult Study (HD-YAS).19 PreHD participants were gene-positive with a CAG repeat >39, Disease Burden Score <24035 and a Unified Huntington’s Disease Rating Scale Total Motor Score of ≤5.36 Control participants were gene-negative family members or individuals with no familial history of Huntington’s disease. Participants were excluded for recent drug or alcohol abuse and/or dependence, neurological or significant psychiatric comorbidity, brain trauma or contraindication to MRI. All participants underwent an extensive battery of cognitive and neuropsychiatric testing, clinical and medical history, neuroimaging, blood sampling and optional CSF collection.19 The study was approved by the local Research Ethics Committee and all participants gave written informed consent prior to study entry.
Biofluid collection
Biofluids were acquired using standardized, validated conditions, methods and equipment.37 The NfL protein32,33 was collected from both CSF and blood plasma.
MRI data acquisition
MRI data were acquired on a 3 T Prisma Scanner (Siemens Healthcare) with a 64-channel head coil. T1-weighted images were acquired using a 3D magnetization prepared rapid gradient echo (MPRAGE) sequence: repetition time (TR) = 2530 ms; echo time (TE) = 3.34 ms; inversion time (TI) = 1100 ms; flip angle = 7°; field of view = 256 × 256 × 176 mm3 with a resolution of 1.0 × 1.0 × 1.0 mm3. Diffusion weighted images were acquired using a multiband spin-echo echo planar imaging (EPI) sequence with TR = 3260 ms, TE = 58 ms, flip angle = 88°, field of view = 220 × 220 mm2. Seventy-two slices were collected with a resolution of 2 × 2 × 2 mm3. The multi-shell data consisted of b-values of 0 (n = 10, one with reverse phase-encoding), 100 (n = 8), 300 (n = 8), 1000 (n = 64) and 2000 (n = 64) s/mm2. Blip reversal acquisition parameters (used in topup) were the same as above. Resting-state functional MRI (fMRI) data were collected using a standard 2D EPI sequence: TR = 3.36 s; TE = 30 ms; 48 slices were acquired with 2.5 mm slice thickness with in-plane field of view of 192 × 192 mm2 with 3 × 3 mm2 resolution with 165 volumes. Field maps were collected to correct for inhomogeneity in the B0 field of the EPI fMRI images: TR = 1020 ms; TE1 = 10 ms; TE2 = 12.46 ms, 64 slices were acquired with 2 mm slice thickness with in-plane field of view of 192 × 192 mm2 with 3 × 3 mm2 resolution. Pulsatile information was collected using the Nonin 8600FO pulse-oximeter and a Siemens breathing belt for respiratory data. Both were recorded along with scanner pulses using Cambridge Electronics Device CED Micro 1401 Mk II connected to a laptop running Spike v2.
MRI atlases
Cortical and subcortical atlases were derived from fMRI datasets and represent regions parcellated on this basis of their functional connections. For cortical regions, we used the Shaeffer cortical atlas,38 and to investigate the effects of parcellation granularity, we performed NBS and genetic analyses using both the 100 and 500 region parcellations. For striatal regions, we used the Choi atlas39 generated by assigning each voxel in the striatum to the most strongly correlated cortical region on the basis of its functional connectivity. Both atlases were registered to standard Montreal Neurological Institute space and then combined into one atlas, resulting in 114 and 514 region atlases.
Diffusion processing
A white matter connectome was created for each participant using anatomically constrained tractography40 implemented in MRtrix.41 Raw diffusion images were first visually quality controlled. Denoising42 and Gibbs ringing artefact removal was performed43 using MRtrix. FSL eddy and topup were used to correct image distortions due to eddy current- and susceptibility-induced off-resonance fields and subject movement.44 B1 field inhomogeneity correction for the diffusion weighted imaging volume series was then performed using the ANTS N4 algorithm.45 Voxel-wise fibre orientation distribution was calculated using multi-shell multi-tissue constrained spherical deconvolution,46 with group-averaged response functions estimated for white matter, grey matter and CSF. Intensity normalization was then performed on fibre orientation distributions and probabilistic whole-brain tractography implemented to generate 10 million streamlines. Streamlines terminated when exiting the white matter. Spherical deconvolution informed filtering of tractograms (SIFT2) was used to remove biases inherent in tractography where longer connections are overdetermined, streamlines follow the straightest path and lack an associated volume.47 Connectomes were constructed by combining streamline tractograms with each participant’s combined cortical (100 and 500 regions of interest)/subcortical (14 regions of interest) parcellation and streamlines assigned to the closest region within a 2 mm radius of each end point. Structural connections were then weighted by streamline count and a cross-sectional area multiplier as per SIFT2.48 Connections were then combined into 114 × 114 and 514 × 514 undirected and weighted matrices.
Functional MRI processing
Functional MRI data preprocessing and subsequent statistical analyses were performed using SPM12 running under MATLAB (ver R.2012b). The T1-weighted scan was segmented into grey and white matter during this process and DARTEL deformation parameters were created. The first five EPI images were discarded to allow for steady-state equilibrium. Functional images were slice-timing corrected and realigned, incorporating field maps for inhomogeneity correction, and coregistered to the T1 image. EPI images were then normalized using DARTEL deformation parameters and smoothed using a 6 mm full-width at half-maximum Gaussian kernel. Functional connectivity analyses were then performed using the CONN toolbox.49 Smoothed, normalized EPI images were included with corresponding structural images (combined, segmented grey and white matter). All EPI images were denoised using a bandpass filter 0.008–0.09 and linear detrending, movement parameters and signals from both white matter and CSF as a proxy for physiological measures were additionally regressed. Regression was performed before bandpass filtering, as is the default in the CONN toolbox. This avoids the reintroduction of motion artefacts50 or unwanted frequency components,51 which can occur when regression is performed after bandpass filtering. Connections were then combined into 114 × 114 and 514 × 514 undirected and weighted matrices, matching the structural connectivity matrices. This approach has been used both by our group11 and others.52 Simultaneous50,51 regression and bandpass filtering were also performed using the ‘simult’ option in CONN.
Motion parameters in six directions were derived for each individual following the realignment step that was performed as part of the fMRI data preprocessing pipeline. These motion parameters were subsequently included as a covariate of no interest in the first-level analyses for each participant. The motion-corrected data were then used at the second level with potential differences due to motion essentially removed. It should also be noted that while this was a movement-disordered patient group, all participants were asymptomatic and as such there was minimal effect of disease-related motion on the scans, each of which were quality-controlled prior to any preprocessing or analyses. Additionally, the maximum movement displacement was calculated for each subject and group differences were explored using a two-tailed t-test.
Connectivity analyses
NBS version 1.2 (https://sites.google.com/site/bctnet/comparison/nbs) was used to investigate independently group differences in structural and functional connectivity53 using both the 114 and 514 parcellation matrices. Using this method, a test statistic is calculated for each connection independently. A primary threshold (P < 0.05, uncorrected) is then applied to form a set of suprathreshold connections. Permutation testing is then used to calculate a family-wise error (FWE) corrected P-value for each set of suprathreshold connections or subnetwork.53 Results reaching FWE corrected P < 0.05 are reported as significant, with P-values relating to the significance of all the connections within a subnetwork as a whole as opposed to individual connections. For these analyses, permutation testing using unpaired t-tests and 5000 permutations, as per the default NBS options, was performed on a general linear model that included age and gender as covariates. A test statistic was then computed for each connection and a default threshold applied (t = 3.1) to produce a set of suprathreshold connections that displayed significant between-group connectivity differences. FWE-correction was applied at P = 0.05.
To focus only on functional connections that have an underlying structural connection, the functional connectivity analysis was repeated constraining the functional connectome by the structural. Here, the functional matrix was simply multiplied by the binarized structural matrix to remove any functional connections that do not have supporting structural connections, and the NBS analysis repeated. Statistically significant group differences in connectome density, as defined by the sum of all weighted connections, were also investigated. This was performed for structural, functional and constrained connectomes using permutation testing (5000 permutations) with two-tailed t-tests including age and gender as covariates.
The relationship between NfL and connectivity may occur in a continuous manner such that higher NfL levels correlate with absent or reduced connectivity. Alternatively, connectivity changes may occur (or be detectable) only when a certain pathological threshold of NfL is reached. To test these two hypotheses, we performed two sets of NfL analyses.
First, we investigated the role of CSF NfL on structural and functional connectivity, correlating CSF NfL to structural and functional connections by including CSF NfL as the contrast in the NBS design matrix for the whole cohort, preHD only and control only.
Next, to investigate whether preHD participants with CSF NfL above a pathological threshold showed differences in structural or functional connectivity compared to preHD participants with normal CSF NfL, a subgroup analysis was performed where the preHD group was split in two on the basis of the CSF NfL results in the study. The low group had CSF NfL values within the 95th percentile of controls (<951 pg/ml), whereas the high group had CSF NfL values above this. This resulted in 24 gene carriers in the low group and 22 in the high group. The 95th percentile of controls was defined as the pathological threshold, in keeping with previous analyses using this cohort.19
Gene expression analysis
Mapping gene expression data to MRI space
Gene expression microarray data were sourced from the Allen Human Brain Atlas (AHBA)54 to examine gene expression underlying the relationship between NfL and functional connectivity, as we identified significant association in our primary connectivity analyses. This contains gene expression data of 20 737 genes sampled across the adult brain. This atlas is based on data from six post-mortem human brains with no known neuropsychiatric or neuropathological history. Five donors were male and one was female with a mean age of 42.5 years. Three were Caucasian, two were African-American and one was Hispanic. AHBA data are freely available to download from the Allen Institute of Brain Science (AIBS, http://human.brain-map.org/static/download). The Abagen toolbox (https://github.com/rmarkello/abagen) was used to map gene expression data on to the combined cortex and striatum 114 regions of interest atlas. This toolbox follows optimized preprocessing steps previously reported.55 In brief, each tissue sample was assigned to one of the 114 regions of interest using AHBA MRI data for each donor. Data were pooled between homologous cortical regions (to ensure adequate bi-hemispheric coverage), with a 2 mm distance threshold on the cortical surface between samples. Probes with expression measures above background in over 50% of samples were selected, and a representative probe per gene was chosen based on highest intensity. Gene expression data were then normalized, leading to 15 633 genes included in the final gene dataset. In the AHBA, data for the left hemisphere were available for all donors, while two donors included right hemisphere data. Previous studies have used mirroring, were the left hemisphere data are mirrored on the right hemisphere in order to account for this.56 We opted not to perform mirroring, as this approach has a differential impact on statistical estimates in regional gene expression analyses.57
Statistical analysis: partial least squares regression
All statistical analysis was performed in MATLAB R2018b. Partial least squares (PLS) regression was used to reveal the biological and cell-specific mechanisms underlying the relationship between CSF NfL and functional connectivity. PLS regression is a multivariate technique used to identify associations between response and predictor variables. In our case, the predictor variable was a 114 regions of interest × 15 633 gene matrix.
Two complimentary approaches were used to generate the response variable, a partial correlation analyses in the preHD group only and a mixed linear model with a focus on the NfL × Group interaction. For the partial correlation analysis, graph theory strength was calculated, which equates to the sum of functional connectivity for each region of interest. Spearman rank partial correlations were then performed with NfL, controlling for age and gender. The partial correlations for each region of interest were then used as the response variable for the PLS. In this context, the focus is not on which correlations are significant but rather the spectrum of correlations across cortical regions of interest; similar approaches have been used in the literature to relate age, cortical thickness and regional gene expression.58 Strength was selected as a graph theory metric, as it is calculated by the sum of weighted connections to each region of interest. This allows comparison with the NBS analysis, which uses weighted connections as an input in the form of a connectivity matrix. While there is no current consensus within the literature as to the optimal graph theory metric for use in resting-state fMRI analyses, graph theory strength has higher test–retest reliability, as measured by interclass correlation coefficient, than other commonly used metrics such as clustering coefficient, betweenness centrality, local efficiency and degree.59
To explore the interaction between NfL and group, the following mixed linear model was used: region of interest functional connectivity strength ∼1 + Age + Gender + Group × NfL. The Group × NfL estimate for each region of interest was used as the response variable for the PLS. A mixed linear model was used as this is the most appropriate approach when including dependent variables, such as Group and NfL. In this context the focus is not on which model estimates are significant but rather the spectrum of negative and positive estimates across cortical regions of interest; similar approaches have been used in the literature to relate age, cortical thickness, magnetization transfer ratio and regional gene expression.60
Partial correlations, model estimates and spatial patterns of the weights of the PLS components were visualized using the BrainNet viewer (https://www.nitrc.org/projects/bnv) for combined cortical and subcortical visualizations and ggseg (https://github.com/ggseg/ggsegSchaefer) for visualizations of cortical surface only.
We performed spatial permutation testing to assess whether PLS results explained a significantly higher proportion of variance for each of our chosen response variables (partial correlation in the preHD group and group × NFL interaction, assessed separately) than expected by chance. To do this, we reordered the predictor matrix in terms of regions of interest based on sphere rotations61 and repeated the PLS regression using this predictor variable; this process was repeated for 1000 random permutations to construct a spatially correlated null distribution of PLS weights in keeping with the literature.29,61,62P-values for PLS components have been calculated based on the explained variance in the observed data relative to the variance explained in the null model. To quantify the spatial topography of PLS weights and enable comparison between the 114 and 514 region of interest atlases, Spearman rank correlations were performed between the MRI coordinates (left to right, A: posterior to anterior, S: inferior to superior) and PLS weights, for the 114 and 514 region of interest NfL partial correlation analyses.
As the greatest amount of variance was explained by the first PLS component (PLS1), genes were ranked based on their contribution to this component. Permutation testing was used to assess whether genes were weighted higher or lower than expected by chance, correcting for FWE. Similar to the NBS implementation, the one-sided FWE-corrected P-value (q-value) for a gene is estimated as the proportion of permutations for which the weighting of this gene is higher than the 95th percentile or lower than the 5th percentile of the spatially correlated null distribution. Only genes with weights significantly higher or lower than expected by chance (q < 0.05) were included in the subsequent GO enrichment analysis. Genes with negative (downweighted) and positive (upweighted) PLS weightings were ranked separately. There are several previous studies that used PLS for the large gene-expression datasets from the AHBA.18,29,63
Gene ontology enrichment analysis
To investigate the genetic basis underlying the CSF NfL and functional connectivity associations, we performed enrichment analysis for GO, Kyoto Encyclopedia of Genes and Genomes pathway, Reactome and CORUM terms using g:Profiler to identify GO terms that were significantly enriched in the top (upweighted) and bottom (downweighted) genes from the PLS1 ranked gene list. Only genes that were significantly more up- or downweighted than expected by chance (against a spatially correlated null distribution) were included in this analysis. A Benjamini–Hochberg correction for multiple comparisons was used with a significance threshold of 0.05, as implemented in g:Profiler. To aid interpretation, we removed general GO terms by excluding those with greater than 1000 genes in their classification in keeping with other studies in the literature.63,64 This allowed us to focus on specific gene sets as opposed to GO terms encompassing thousands of genes covering a range of processes.
Expression-weighted cell-type enrichment analysis
To investigate whether specific cell types were associated with CSF NfL and functional connectivity, we performed EWCE.65 The top (upweighted) and bottom (downweighted) 10%, 20% and 30% of genes from the PLS1 ranked gene list were used as target lists. Incremental thresholds were chosen to identify the most significant cell-type association for each target list. Each was run with 100 000 bootstrap lists, controlling for transcript length and GC content, which can bias genetic enrichment analyses,66 using only major cell-type classes (e.g. ‘astrocyte’, ‘microglia’, etc.). The Benjamini–Hochberg method was used for correction of multiple comparisons as is the default in EWCE software. Single-cell transcription data were used from the AHBA (https://portal.brain-map.org/atlases-and-data/rnaseq) containing data from the middle temporal gyrus.67 To ensure that our results were not dependent on the dataset used, we replicated our EWCE analysis with the same parameters (100 000 bootstrap lists, Benjamini–Hochberg correction) using a different human-derived dataset from Habib et al.68; this is a comprehensive human derived post-mortem dataset, containing data from five donors and 19 550 cells from both the hippocampus and the prefrontal cortex. The EWCE package is freely available from https://github.com/NathanSkene/EWCE.
Enrichment analysis of striatal and cortical genes showing abnormal transcription in Huntington’s disease
We then investigated whether striatal and cortical genes showing abnormal transcription in human and animal models of Huntington’s disease were enriched greater than by chance in the ranked gene list of the PLS1. Huntington’s disease gene lists were obtained from Langfelder et al.,69 which consists of genes that show consistent differences in Huntington’s disease compared to controls both in the Huntington’s disease knockout mouse allelic series69 and human Huntington’s disease post-mortem data from the caudate nucleus70 and cortical regions Brodmann areas 4 and 9,31 prefrontal and visual cortices.71 To test whether these gene lists were enriched greater than by chance in the PLS1, we performed a permutation test of the normalized bootstrap weight of each gene in the PLS1 summed over all genes for each gene list. The approach has been used previously,18,63 and the code is freely available at https://github.com/KirstieJane/NSPN_WhitakerVertes_PNAS2016/blob/master/SCRIPTS/PLS_candidate_genes.m.
Enrichment analysis of cell-specific genes showing abnormal transcription in snRNAseq in Huntington’s disease
To relate cell-specific CSF NfL–fMRI relationships to Huntington’s disease pathology, we utilized data from a study analysing snRNAseq data in post-mortem Huntington’s disease brains relative to controls.72 Single nucleus RNAseq can be applied to frozen post-mortem brain tissue and thus overcomes limitations of single-cell (sc)RNAseq approaches, which cannot be applied to frozen tissue. This enables the identification of cell-specific genes that show abnormal transcription in Huntington’s disease. Al-Dalahmah et al.72 analysed snRNA seq data from samples of the anterior cingulate cortex frozen at post-mortem in four cases (two Huntington’s disease and two controls) from the New York Brain Bank. In doing so, they provided lists of neuron- and astrocyte-specific genes that signify different levels of transcription in Huntington’s disease relative to controls. We tested whether these gene lists were enriched greater than by chance in the PLS1 from the above gene CSF NfL-functional connectivity analysis using the permutation test described above. See Fig. 1 for a summary of the methodical approach.
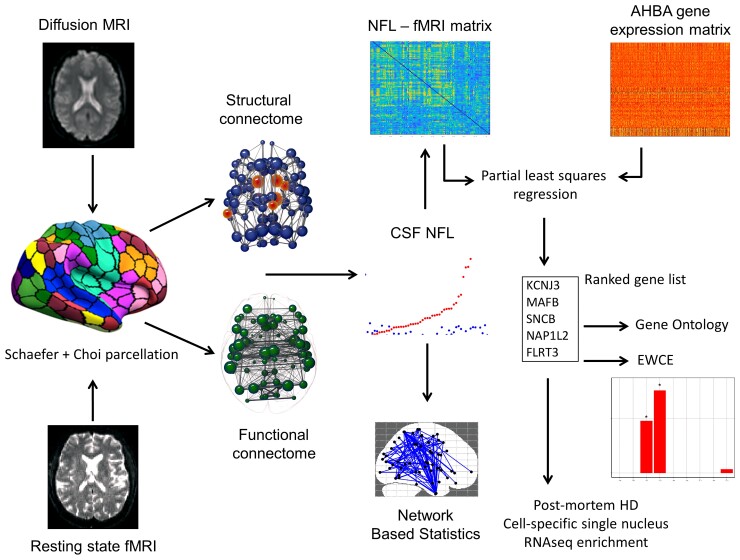
Figure 1.
Summary of analysis pipeline. Diffusion MRI and resting-state fMRI underwent preprocessing and were parcellated using the Shaefer cortical atlas38 and the Choi subcortical atlas.39 Structural and functional connectomes were then created based on weighted streamlines between brain regions and temporal fMRI time-series correlations between regions, respectively. Correlations were performed between CSF NfL and brain networks using NBS. An NfL–fMRI correlation matrix was also used to investigate associations with regional gene expression using the AHBA. PLS regression produced a ranked gene list of those genes most strongly associated with NfL–fMRI hyperconnectivity. GO and EWCE were then used to investigate biological and cell-specific associations. Finally these results were validated using snRNAseq72 from post-mortem Huntington’s disease (HD) and control brains.
Data and code availability
Anonymized, derived data supporting the findings of this study are available from the corresponding author on request. Code used to implement analyses in this study is freely available at https://github.com/AngelikaZa/YAS_HD.
Results
Demographic and clinical data
There were no significant differences in age [t(85) = 0.7, P = 0.49] between controls (mean = 28.61, SD = 5.68) and gene carriers (mean = 29.46, SD = 5.62), sex (χ2, P = 0.67) between controls (females = 23, males = 18) and gene carriers (female = 23, male = 23) or International Standard Classification of Education (χ2, P = 0.45). However, CSF NfL levels were significantly different [t(85) = 4.2, P = 0.0001] between controls (mean = 354, SD = 261) and gene carriers (mean = 767, SD = 585).
Structural and functional connectivity
There were no significant between-group differences in structural or functional connectivity (with and without constraining by structural connectome) for either the 114 or 514 parcellation analyses (Table 1). No connectome density group differences were observed for structural (114 regions of interest, P = 0.52; 514 regions of interest, P = 0.66), functional (114 regions of interest, P = 0.65; 514 regions of interest, P = 0.64) or constrained connectomes (114 regions of interest, P = 0.58; 514 regions of interest, P = 0.83). For resting-state fMRI, there was no significant difference (P = 0.096) in maximum movement displacement between preHD (mean = 0.75, SD = 0.49) and controls (mean = 0.62, SD = 0.24).
Table 1.
Network-based statistics results
| Group t-test | PreHD < Controls | Cont < PreHD |
|---|---|---|
| Functional (114 ROIs) | 0.672 | 0.6046 |
| Structural (114 ROIsr) | 0.3447 | 0.2384 |
| Functional (structural constrained) 114 ROIsr | 0.6072 | 0.5792 |
| Functional (514 ROIsr) | 0.4296 | 0.5976 |
| Structural (514 ROIsr) | 0.1504 | 0.4613 |
| Functional (structural constrained) 514 ROIsr | 0.2603 | 0.7998 |
| Correlations | Positive | Negative |
| NFL–fMRI correlation (whole group) 114 ROIsr | 0.0304* | 1 |
| NFL–fMRI correlation (preHD) 114 ROIsr | 0.019* | 0.6633 |
| NFL–structural correlation (whole group) 114 ROIsr | 0.191 | 0.391 |
| NFL–structural correlation (preHD) 114 ROIsr | 0.2523 | 0.1818 |
| NFL–fMRI correlation (whole group) 514 ROIsr | 0.0398* | 0.6615 |
| NFL–fMRI correlation (preHD) 514 ROIsr | 0.027* | 0.6478 |
| NFL–structural correlation (whole group) 514 ROIsr | 0.3177 | 0.0284* |
| NFL–structural correlation (preHD) 514 ROIsr | 0.2919 | 0.023* |
For these analyses, permutation testing using unpaired t-tests and 5000 permutations was performed on a general linear model that included age and sex as covariates. A test statistic was then computed for each connection and a default threshold applied (t = 3.1) to produce a set of suprathreshold connections that displayed significant between-group connectivity differences. *FWE-correction was applied at P = 0.05. Due to the high false positive rates in fMRI connectivity analyses73 the functional connectivity analysis was repeated, constraining the functional connectome by the structural. Here, the functional matrix was simply multiplied by the structural matrix to remove any functional connections that do not have supporting structural connections, and the NBS analysis repeated. ROIs = regions of interest.
Relationship between CSF NfL and structural and functional connectivity in preHD
A three-way ANOVA was performed to compare structural and functional connectivity using 114 parcellations between three groups: controls, preHD with normal CSF NfL and preHD with higher CSF NfL. No significant differences were found for either structural (PFWE = 0.25) or functional (PFWE = 0.97) connectivity.
For structural connectivity, there were no significant correlations between CSF NfL for controls and preHD combined or preHD only for the 114 parcellation. There were significant negative correlations for both controls and preHD combined (PFWE = 0.028) and preHD only (PFWE = 0.023) for the 514 parcellation (Supplementary Figs 1 and 2).
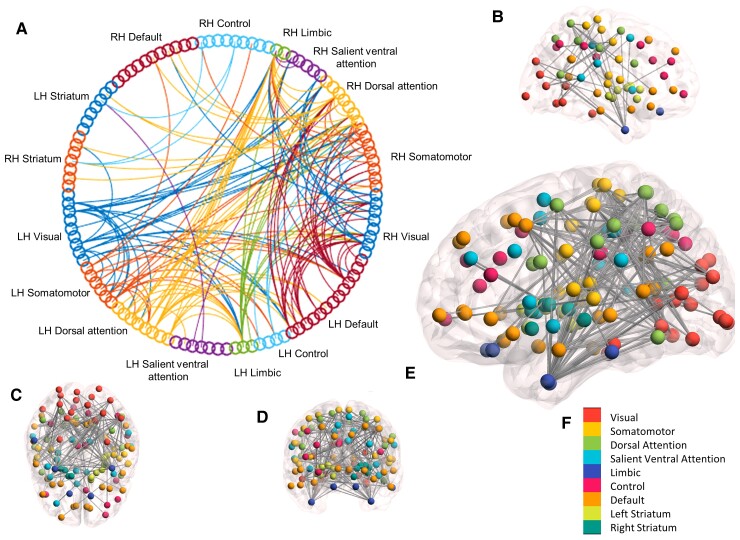
For functional connectivity, there were significant positive correlations between CSF NfL and functional connectivity for both the combined (PFWE = 0.034) and preHD group only (PFWE = 0.019) for the 114 parcellation (Table 1 and Fig. 2) and for both the combined (PFWE = 0.04) and preHD group only (PFWE = 0.027) for the 514 parcellation (Supplementary Figs 1 and 2). The connections of the subnetwork that showed a positive correlation with NfL were located predominantly in the posterior cortex, with very few anterior regions affected. Of the 164 connections in the preHD group-only subnetwork, 6% were cortico-striatal, 49% interhemispheric and 45% intrahemispheric (Table 2 and Supplementary Table 2). There were no significant negative correlations between CSF NfL and functional connectivity (Table 1).
Figure 2.
Resting-state fMRI brain subnetwork showing significant (P < 0.05 FWE-corrected) positive correlation with CSF NfL across Huntington’s disease gene expansion carriers. Analysis performed using NBS. (A) Circular graph depicting significant subnetwork. (B) Left sagittal view. (C) Axial view. (D) Coronal view. (E) Right sagittal. (F) Colour scheme for brain figures. Spheres indicate brain regions; lines indicate fMRI connections that correlate with NfL between brain regions. LH = left hemisphere; RH = right hemisphere.
Table 2.
Network-based statistics subnetwork showing significant correlation with CSF NfL for preHD gene carriers
| Connection 1 | Connection 2 | T-stat | |
|---|---|---|---|
| Cortical–striatal | 7Networks_RH_Cont_PFCl_3 | R_Ventral_attention | 4.06 |
| 7Networks_LH_SalVentAttn_PFCl_1 | L_Dorsa_lattention | 3.95 | |
| 7Networks_RH_DorsAttn_Post_4 | R_Somatomotor | 3.88 | |
| 7Networks_RH_Cont_pCun_1 | R_Ventral_attention | 3.51 | |
| 7Networks_RH_DorsAttn_Post_4 | L_Somatomotor | 3.44 | |
| 7Networks_RH_DorsAttn_Post_1 | L_Somatomotor | 3.42 | |
| 7Networks_RH_Cont_PFCl_3 | L_Dorsal_attention | 3.27 | |
| 7Networks_RH_DorsAttn_Post_5 | R_Frontoparietal | 3.24 | |
| 7Networks_LH_Vis_7 | R_Somatomotor | 3.18 | |
| 7Networks_RH_DorsAttn_Post_4 | R_Ventral_attention | 3.15 | |
| Interhemispheric | 7Networks_LH_Default_Temp_1 | 7Networks_RH_DorsAttn_Post_5 | 5.24 |
| 7Networks_LH_Cont_pCun_1 | 7Networks_RH_SomMot_6 | 5.13 | |
| 7Networks_LH_DorsAttn_Post_5 | 7Networks_RH_Limbic_TempPole_1 | 4.9 | |
| 7Networks_LH_Default_Par_1 | 7Networks_RH_Vis_2 | 4.79 | |
| 7Networks_LH_Cont_pCun_1 | 7Networks_RH_SomMot_2 | 4.76 | |
| 7Networks_LH_DorsAttn_FEF_1 | 7Networks_RH_Limbic_TempPole_1 | 4.74 | |
| 7Networks_LH_Default_PFC_1 | 7Networks_RH_DorsAttn_Post_5 | 4.71 | |
| 7Networks_LH_Limbic_TempPole_1 | 7Networks_RH_DorsAttn_Post_5 | 4.65 | |
| 7Networks_LH_Cont_pCun_1 | 7Networks_RH_SomMot_5 | 4.64 | |
| 7Networks_LH_SomMot_6 | 7Networks_RH_Limbic_TempPole_1 | 4.58 | |
| Intrahemispheric | 7Networks_RH_SomMot_6 | 7Networks_RH_DorsAttn_Post_1 | 6.78 |
| 7Networks_LH_DorsAttn_Post_6 | 7Networks_LH_Default_Temp_1 | 4.86 | |
| 7Networks_RH_Vis_2 | 7Networks_RH_SomMot_8 | 4.84 | |
| 7Networks_LH_Vis_7 | 7Networks_LH_Limbic_TempPole_1 | 4.64 | |
| 7Networks_RH_DorsAttn_Post_5 | 7Networks_RH_Limbic_TempPole_1 | 4.53 | |
| 7Networks_RH_SomMot_8 | 7Networks_RH_DorsAttn_Post_1 | 4.51 | |
| 7Networks_LH_DorsAttn_Post_6 | 7Networks_LH_Limbic_TempPole_1 | 4.5 | |
| 7Networks_LH_DorsAttn_Post_3 | 7Networks_LH_Limbic_TempPole_1 | 4.49 | |
| 7Networks_LH_SomMot_6 | 7Networks_LH_Default_PFC_6 | 4.41 | |
| 7Networks_LH_Vis_3 | 7Networks_LH_Limbic_TempPole_1 | 4.39 |
Connections classified as cortico-striatal, interhemispheric, intrahemispheric and ranked based on test statistic. Top 10 connections based on test statistic (T-stat) displayed for each connection type.
No significant correlations were seen between NfL and either structural or functional connectivity for the control group only (Supplementary Table 1), suggesting the absence of a physiological relationship between NfL and brain networks. Replication of the significant fMRI analyses using the CONN ‘simult’ processing option did not reveal significance and may be related to less specificity with this processing option (Supplementary Table 1).
Region of interest partial correlation, mixed linear model and PLS analyses
For the partial correlation of 114 regions of interest analysis, one brain region showed false discovery rate (FDR)-corrected significance (q), left DorsAttn_Post_6 (rho = 0.38, q = 0.031). A further seven nodes from the dorsal attention and visual networks showed uncorrected (P < 0.05) significance, all with positive correlations. See Supplementary Table 3 and Supplementary Figs 3–5 for visualizations of correlations. Using the region of interest correlations as an input into the PLS analysis, the first component explained the largest amount of variance at 22.5%, P < 1 × 10−10 [2nd, 8% (P = 0.49); 3rd, 15.4% (P = 0.005); 4th, 10.1% (P = 0.24); 5th, 9% (P = 0.36)]. The spatial patterns of the weights of the PLS1 are visualized in Supplementary Figs 6–8. The spatial topography analysis revealed correlations between PLS weights and R (left to right): rho = 0.02, P = 0.83, A (posterior to anterior): rho = −0.35, P = 0.0004, S (inferior to superior): rho = 0.42, P = 1.3 × 10−5.
For the mixed linear model analysis uncorrected significance for the group × NfL interaction was seen for right Default_pCunPCC_2 (β = −0.008, P = 0.013), right Limbic_TempPole_1 (β = 0.008, P = 0.027) and right Vis_7 (β = 0.01, P = 0.031). See Supplementary Table 4 and Supplementary Figs 9–11 for visualizations of model estimates. Using the region of interest Group × NfL interaction estimate as an input into the PLS analysis, the first component explained the largest amount of variance at 24.7%, P = 0.047 [2nd, 6.1% (P = 0.29); 3rd, 8.9% (P = 0.41); 4th, 8.6% (P = 0.4); 5th, 10.1% (0.47)]. The spatial patterns of the weights for the PLS1 are visualized in Supplementary Figs 12 and 14.
For the partial correlation of 514 regions of interest analysis, no brain regions showed FDR-corrected significance. Ninety-seven nodes from the dorsal attention, visual, somatomotor, limbic and default mode, salient ventral attention and control networks showed uncorrected (P < 0.05) significance (91 positive correlations and six negative correlations). Consistent with the 114 regions of interest analysis, dorsal attention and visual regions of interest were among the most significant. See Supplementary Table 5 and Supplementary Fig. 15 for visualizations of correlations.
Using the region of interest correlations as an input into the PLS analysis, the first component explained the largest amount of variance at 13.03%, P = 0.014. Downweighted but no upweighted genes were identified in the first component; therefore, the second component was also investigated in subsequent analyses. The spatial pattern of the weights for the first PLS component are visualized in Supplementary Fig. 16. The spatial topography analysis revealed correlations between PLS weights and R (left to right): rho = 0.18, P = 5.5 × 10−5, A (posterior to anterior): rho = −0.42, P = 2.2 × 10−16, S (inferior to superior): rho = 0.43, P = 2.2 × 10−16.
The second component explained 9.14% of the variance, P = 0.014 [3rd, 2.98% (P = 0.41); 4th, 7.92% (P = 0.036); 5th, 5.86% (P = 0.12)]. The spatial pattern of the weights for the second PLS component (PLS2) are visualized in Supplementary Fig. 17. The spatial topography analysis revealed correlations between PLS weights and R (left to right): rho = −0.21, P = 1.2 × 10−6, A (posterior to anterior): rho = −0.35, P = 5.8 × 10−16, S (inferior to superior): rho = 0.23, P = 2.2 × 10−7. Based on spatial topography, correlations for both 114 and 514 regions of interest analyses showed higher PLS weights in posterior and superior cortical regions of interest (Supplementary Figs 18–20). To facilitate comparisons between the 114 and 514 regions of interest across NBS, GO, EWCE and Huntington’s disease gene enrichment analyses, Supplementary Table 13 summarizes the results across the analyses.
Gene ontology enrichment analysis
For the results using the ranked gene list from the partial correlation of 114 regions of interest PLS, the five most significant ontology terms for upweighted genes included presynapse (P = 4.84 × 10−9), somatodendritic compartment (P = 6.85 × 10−9), synaptic membrane (P = 1.75 × 10−9), potassium ion transmembrane transporter activity (P = 2.11 × 10−8) and presynaptic membrane (P = 3.93 × 10−8) (see Table 3). For downweighted genes, the five most significant ontology terms included cell morphogenesis involved in differentiation (P = 0.0003), I band (P = 0.003), phosphatidylinositol-4,5-bisphosphate binding (P = 0.004), camera-type eye development (P = 0.006) and cell morphogenesis involved in neuron differentiation (P = 0.007) (see Table 3).
Table 3.
Top 5 significant GO terms for upweighted and downweighted genes from the partial correlation analysis (Pcor) and the mixed linear model (Group × NfL) interaction analysis
| Term name | Term ID | Source | P-value |
|---|---|---|---|
| Upweighted (Pcor) | |||
| Presynapse | GO:0098793 | GO:CC | 4.84 × 10−9 |
| Somatodendritic compartment | GO:0036477 | GO:CC | 6.85 × 10−9 |
| Synaptic membrane | GO:0097060 | GO:CC | 1.75 × 10−8 |
| Potassium ion transmembrane transporter activity | GO:0015079 | GO:MF | 2.11 × 10−8 |
| Presynaptic membrane | GO:0042734 | GO:CC | 3.93 × 10−8 |
| Downweighted (Pcor) | |||
| Cell morphogenesis involved in differentiation | GO:0000904 | GO:BP | 0.0002733 |
| I band | GO:0031674 | GO:CC | 0.0028901 |
| Phosphatidylinositol-4,5-bisphosphate binding | GO:0005546 | GO:MF | 0.0036894 |
| Camera-type eye development | GO:0043010 | GO:BP | 0.0055427 |
| Cell morphogenesis involved in neuron differentiation | GO:0048667 | GO:BP | 0.0066415 |
| Upweighted (Group × NfL) | |||
| Microtubule organizing centre | GO:0005815 | GO:CC | 0.0008331 |
| Plasma membrane-bounded cell projection assembly | GO:0120031 | GO:BP | 0.0012018 |
| Cell projection assembly | GO:0030031 | GO:BP | 0.0018766 |
| Centrosome | GO:0005813 | GO:CC | 0.0029856 |
| Cilium organization | GO:0044782 | GO:BP | 0.0040246 |
| Downweighted (Group × NfL) | |||
| Presynapse | GO:0098793 | GO:CC | 2.13 × 10−14 |
| Axon | GO:0030424 | GO:CC | 3.07 × 10−9 |
| Anterograde trans-synaptic signalling | GO:0098916 | GO:BP | 3.31 × 10−9 |
| Chemical synaptic transmission | GO:0007268 | GO:BP | 3.31 × 10−9 |
| Trans-synaptic signalling | GO:0099537 | GO:BP | 5.36 × 10−9 |
GO:BP = gene ontology biological process; GO:CC = gene ontology cellular component; GO:MF = gene ontology molecular function; REAC = reactome.
Results using the ranked gene list from the Group × NfL interaction for downweighted genes were similar were similar to the upweighted ontology terms for the partial correlation analysis. The five most significant ontology terms for downweighted genes included presynapse (P = 2.13 × 10−14), axon (P = 3.07 × 10−9), anterograde trans-synaptic signalling (P = 3.31 × 10−9), chemical synaptic transmission (P = 3.31 × 10−9) and trans-synaptic signalling (P = 5.36 × 10−9). The five most significant ontology terms for upweighted genes included microtubule organizing centre (P = 0.0008), plasma membrane-bounded cell projection assembly (P = 0.001), cell projection assembly (P = 0.002), centrosome (P = 0.003) and cilium organization (P = 0.004) (see Table 3).
For the results using the ranked gene list from the partial correlation of 514 regions of interest PLS, for the PLS1, there were no upweighted genes; therefore, we included upweighted genes from the second component. Significant ontology terms for upweighted genes (component 2) included overlap with terms reported in for upweighted genes in the partial correlation of 114 regions of interest PLS analyses; these included potassium ion transmembrane transporter activity (P = 1.03 × 10−8), presynapse (P = 6.12 × 10−6) and somatodendritic compartment (P = 9.49 × 10−5). Significant ontology terms for downweighted genes (component 1) included similar synaptic and ion channel gene terms, such as presynapse (9.17 × 10−6), trans-synaptic signalling (P = 2.59 × 10−5) and ion transmembrane transporter activity (P = 1.54 × 10−5). GO lists for the 50 most significant terms, for all analyses, are included in Supplementary Table 6.
Cell-specific enrichment analysis
For the cell enrichment analyses, we focused on the top and bottom 10% of genes. Results were consistent across databases [AIBS 2019 and DRONC (droplet-based single-nucleus RNA sequencing)] and 10–30% gene lists (Supplementary Tables 6 and 12).
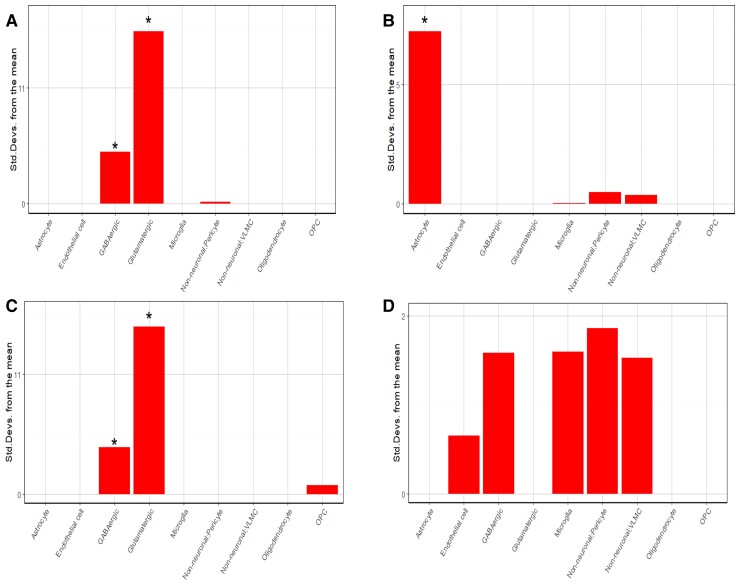
For results using the ranked gene list from the partial correlation of 114 regions of interest analysis, upweighted genes were significantly associated with neuronal cell types, while downweighted genes were significantly associated with glial cell types. For 10% upweighted, AIBS 2019 showed significance for glutamatergic (P < 1 × 10−10) and GABAergic cells (P = 2 × 10−5), while 10% downweighted showed significance for astrocytes (P < 1 × 10−10) (Table 4 and Fig. 3). The neuronal and glial cell split between up- and downweighted genes was replicated using the DRONC database. Full results are included in Supplementary Tables 7 and 8.
Table 4.
EWCE analysis
| Cell type | P-value | Fold change | SD from mean |
|---|---|---|---|
| Upweighted 10% (Pcor) | |||
| Glutamatergic | <1 × 10−10 | 2.89657 | 16.33697 |
| GABAergic | 0.00002 | 1.53571 | 4.95972 |
| Non-neuronal: pericyte | 0.39915 | 1.03437 | 0.20818 |
| OPC | 0.85648 | 0.87782 | −1.04623 |
| Microglia | 0.98576 | 0.71634 | −1.98875 |
| Non-neuronal: VLMC | 0.99380 | 0.57872 | −2.21417 |
| Endothelial cell | 0.99947 | 0.57450 | −2.78966 |
| Astrocyte | 0.99999 | 0.56620 | −3.54350 |
| Oligodendrocyte | 1.00000 | 0.49659 | −3.80922 |
| Downweighted 10% (Pcor) | |||
| Astrocyte | <1 × 10−10 | 1.90801 | 7.21517 |
| Non-neuronal: pericyte | 0.29683 | 1.08003 | 0.49469 |
| Non-neuronal: VLMC | 0.33726 | 1.07361 | 0.37335 |
| Microglia | 0.46827 | 1.00395 | 0.02779 |
| OPC | 0.52067 | 0.98585 | −0.12059 |
| Endothelial cell | 0.80142 | 0.86733 | 0.86733 |
| GABAergic | 0.84056 | 0.89153 | −0.99208 |
| Oligodendrocyte | 0.99660 | 0.69286 | −2.32499 |
| Glutamatergic | 0.99942 | 0.67336 | −2.82282 |
| Upweighted 10% (Group × NfL) | |||
| Non-neuronal: pericyte | 0.03905 | 1.28857 | 1.85843 |
| Microglia | 0.06393 | 1.21547 | 1.59846 |
| GABAergic | 0.06428 | 1.16696 | 1.58190 |
| Non-neuronal: VLMC | 0.06945 | 1.26748 | 1.52874 |
| Endothelial cell | 0.24742 | 1.09724 | 0.65320 |
| Oligodendrocyte | 0.77574 | 0.90052 | −0.78095 |
| Glutamatergic | 0.87605 | 0.87073 | −1.13485 |
| OPC | 0.97856 | 0.79741 | −1.84178 |
| Astrocyte | 0.99049 | 0.74715 | −2.08759 |
| Downweighted 10% (Group × NfL) | |||
| Glutamatergic | <1 × 10−10 | 2.832749 | 15.3434863 |
| GABAergic | 0.0001 | 1.4754347 | 4.27832 |
| OPC | 0.20687 | 1.0899718 | 0.7888829 |
| Non-neuronal: VLMC | 0.98562 | 0.6028899 | −1.9850374 |
| Endothelial cell | 0.98776 | 0.6848202 | −2.026815 |
| Non-neuronal: pericyte | 0.99747 | 0.5892623 | −2.454821 |
| Astrocyte | 0.99978 | 0.6348877 | −2.9696604 |
| Oligodendrocyte | 0.9999 | 0.6004525 | −2.9841701 |
| Microglia | 0.99993 | 0.5594399 | −3.0422093 |
Results for top 10% upweighted and downweighted genes using AHBA 2019 cell-specific gene classification.
OPC = oligodendrocyte precursor cells; Pcor = corrected significance; SD = standard deviation; VLMC = vascular leptomeningeal cells.
Figure 3.
EWCE analysis using AHBA 2019 cell-specific gene annotation. (A) Top 10% upweighted genes for the partial correlation analysis. (B) Top 10% downweighted genes for the partial correlation analysis. (C) Top 10% downweighted genes for the Group × NfL analysis. (D) Top 10% upweighted genes for the Group × NfL analysis. *Corrected significance. OPC = oligodendrocyte precursor cells; VLMC = vascular leptomeningeal cells.
Results using the ranked gene list from the Group × NfL interaction for downweighted genes were similar to the upweighted gene results for the partial correlation analysis. Downweighted genes were significantly associated with neuronal cell types, while upweighted genes were significantly associated with glial cell types. For 10% downweighted, AIBS 2019 showed significance for glutamatergic cells (P < 1 × 10−10) and GABAergic (P = 1 × 10−4), while 10% upweighted showed significance for pericyte (P = 0.04) (Table 4 and Fig. 3). The neuronal and glial cell split between up- and downweighted genes was replicated using the DRONC database (Supplementary Tables 9 and 10).
For results using the ranked gene list from the partial correlation of 514 regions of interest analysis 10% upweighted, AIBS 2019 showed significance for GABAergic (P < 1 × 10−10) and glutamatergic cells (P < 1 × 10−10), while 10% downweighted showed significance for astrocyte (P < 1 × 10−10) and GABAergic cells (P = 0.002). Full results are included in Supplementary Tables 11 and 12.
Enrichment analysis of striatal and cortical genes showing abnormal transcription in Huntington’s disease
For the partial correlation analysis, genes which showed abnormal transcription in the cortex in human Huntington’s disease and animal models were significantly enriched in the ranked gene list from the PLS1 (P < 1 × 10−10). However, genes that showed abnormal transcription in the striatum were not significantly enriched (P = 0.99; Fig. 4). This suggests that CSF NfL-related increases in functional connectivity are predominantly related to cortical and not striatal Huntington’s disease pathology. Neither striatal (P = 0.16) nor cortical genes (P = 0.8) were enriched in the ranked gene list from the PLS1 of the Group × NfL analysis. Consistent with the 114 regions of interest partial correlation analysis, the 514 regions of interest partial correlation analysis showed enrichment of cortex genes (PLS1, P = 0.003; PLS2, P = 2 × 10−4) but not striatal genes (PLS1, P = 0.98; PLS2, P = 0.86).
Figure 4.
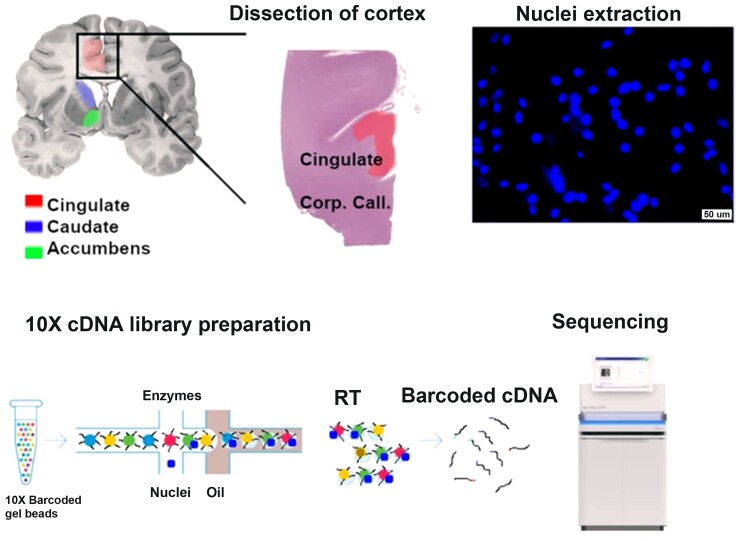
Validation using snRNAseq of the cingulate cortex in control and Huntington’s disease. (A) Experimental scheme from Al-dalahmah et al.72. First, cingulate cortex was dissected, nuclei were extracted and visualized using DAPI nuclear stain under a fluorescence microscope to ascertain membrane integrity. The nuclei were subjected to 10× chromium scRNAseq workflow involving encapsulation of nuclei in oil droplets along with enzymes and barcoded beads, followed by cDNA synthesis and library preparation, and finally, sequencing. (Reproduced from Al-Dalahmah et al.72 under the terms of the Creative Commons CC BY licence.) (B) P-values for analysis testing enrichment of Huntington’s disease striatum and cortex genes from Langfelder et al.69 and Huntington’s disease cell-specific neuronal, astrocyte and microglia genes from Al-Dalahmah et al.72
Enrichment analysis of cell-specific genes showing abnormal transcription in snRNAseq in Huntington’s disease
For the partial correlation analysis, neuronal (P < 1 × 10−10) and microglia (P = 0.03, uncorrected) genes that showed abnormal transcription in Huntington’s disease post-mortem brains were significantly enriched in the ranked gene list from the PLS1. Astrocyte genes abnormally transcribed in Huntington’s disease were not significantly enriched (P = 1), suggesting the cortical pathology associated with CSF NfL functional connectivity increases is associated with neuronal Huntington’s disease-related changes (Fig. 4). Neither neuronal (P = 1), astrocytic genes (P = 0.87) nor microglia (P = 0.97) were enriched in the ranked gene list from the PLS1 of the Group × NfL analysis. Consistent with the 114 regions of interest partial correlation analysis, the 514 regions of interest partial correlation analysis showed enrichment of neuronal genes for PLS2 (P < 1 × 10−10) but not PLS1 (P = 0.09). No significant enrichment was seen for astrocytic or microglia genes in PLS1 1 or PLS2.
Discussion
We characterized functional brain networks in asymptomatic preHD gene carriers very far from disease onset and related these networks to measures of white matter organization, disease burden and gene expression. Despite there being no differences in functional or structural connectivity comparing controls and preHD participants, we identified a significant positive correlation, predominantly in posterior regions, between functional connectivity and disease burden as measured by CSF NfL, a fluid biomarker of axonal degeneration, detectable in those many years from Huntington’s disease clinical diagnosis. Using data from the AHBA and performing cell-enrichment analysis, we demonstrated that those regions that showed increased functional connectivity were also those with regional expression of genes specific to neuronal GABAergic and glutamatergic cells. This relationship was validated using snRNAseq data from post-mortem Huntington’s disease and healthy control brains, where increased functional connectivity was associated with neuronal genes abnormally transcribed in Huntington’s disease.
Studies have shown that functional connectivity differs between preHD and controls5,9,10,12 in cohorts where gene carriers were more advanced, i.e. they showed subtle symptoms and were on average 10–15 years from clinical diagnosis when cognitive changes tend to become evident.74,75 Here, in asymptomatic preHD gene carriers, on average 24 years from disease onset with normal cognitive behaviour,19 we found no functional connectivity differences when compared to controls, even when constrained by the structural connectome. However, we did identify a positive association between functional connectivity and CSF NfL, indicating that connectivity changes relate to disease pathology burden rather than being characteristic of asymptomatic preHD per se.
As there were no differences in white matter organization in our previous analyses19,20 and a limited number of white matter connections showing significant negative correlation with CSF NfL (only for the 514 atlas) in this study, this suggests that large-scale functional changes precede those of microstructure in Huntington’s disease gene carriers furthest from disease onset. It is important to note that as a marker of axonal degeneration, CSF NfL increases indicate some degree of underlying molecular change. The limited change in structural connectivity measures suggests that diffusion-weighted measures lack sensitivity at the very earliest stages of Huntington’s disease and changes in these measures can only be detected after a certain threshold of cumulative change at the molecular level. Nevertheless, with currently feasible in vivo measures, functional connectivity appears to change prior to structural connectivity.
Our findings are consistent with our earlier work focusing exclusively on fronto-striatal connectivity.11 In that earlier work, fronto-striatal connectivity related to cognitive flexibility (posterior regions were not interrogated as part of these analyses) differed in preHD participants, while connections from the striatum to both frontal and posterior cortical regions showed higher connectivity with evidence of compensatory activity to support maintained performance (in review). In the present work, we went beyond our earlier study to now identify positive associations between CSF NfL and functional connectivity in posterior cortical regions. This is of particular interest, given that in our previous work we demonstrated a clear anterior–posterior gradient of functional connectivity upregulation.12 However, this was in gene carriers 10–15 years from clinical diagnosis. Thus, one possibility is that there is a shift in compensatory functional connectivity changes, from posterior to anterior, as pathology becomes more significant in the earliest preHD stages. This should be investigated further in future longitudinal studies.
To understand the basis of the NfL-related increases in functional connectivity that we found, we investigated how brain areas where functional connectivity increased might relate to regional gene expression determined from the AHBA. GO showed an association with biological processes involving synaptic transmission, while EWCE analysis indicated specificity to GABAergic and glutamatergic neuronal cells, which was further supported using independent snRNAseq data from post-Huntington’s disease and healthy control brains. There is significant evidence to suggest that upregulated functional connectivity in neurodegeneration is associated with both glutamate excitotoxicity from pyramidal cells76,77 and loss of GABAergic inhibition from interneurons in both mouse models78 and human cells,79,80 which seems to be located within the cortex rather than the striatum.22,23,78–82 Furthermore, there is a dissociation in terms of the way in which degeneration of cortical interneurons relates to the main presenting symptom in Huntington’s disease. Reduced interneurons in the anterior cingulate cortex, for example, are associated with a predominant mood phenotype, while the primary motor cortex is associated with a motor phenotype.79–81 Interestingly, genes showing abnormal transcription in Huntington’s disease cortex were enriched in our analysis, while those showing abnormal transcription in the striatum were not. These findings, however, must be considered with the caveat that the AHBA gene expression data reflect gene expression determined in participants without any neurological disease; post-mortem brain data for Huntington’s disease gene expansion carriers very far from onset are not currently available.
Gene enrichment results for upweighted genes in the partial correlation analysis were similar to downweighted genes in the NfL × Group interaction analysis. While the direction of effect in the partial correlation analysis is intuitive, such that a positive correlation indicates higher functional connectivity is associated with higher NfL, the interpretation of the NfL × Group interaction is more difficult. Furthermore, the absence of a relationship between NfL and functional connectivity in the control group suggests the possibility that the inclusion of the control group in the model could introduce noise and reduce the pathobiological signal of the preHD NfL relationship. Indeed, this may explain the borderline significance for the PLS1 and absence of enrichment for any gene set in the NfL × Group interaction analysis. This is in contrast to the partial correlation analyses for both 114 and 514 regions of interest, which showed significance for PLS components and enrichment for cortical and neuronal gene sets.
There are some limitations to the current study. There are no gene expression post-mortem brain data in far from onset premanifest Huntington’s disease gene carriers currently available. Here we show that the spatial distribution of NfL–functional connectivity correlations are associated with neuronal genes implicated in Huntington’s disease pathogenesis. The manifest Huntington’s disease post-mortem data are used to demonstrate that neuronal genes that show differential expression in post-mortem Huntington’s disease brains relative to controls are enriched in the ranked gene list from our PLS analysis. However, we postulate that while the underlying pathobiology of Huntington’s disease remains consistent across the lifetime of the disease, how this emerges at the brain network levels differs across the disease spectrum; for example, while functional connectivity in far-from-onset gene carriers may increase in the context of increasing disease burden, this may then reduce once a critical level of pathology is reached, such that hyperexcitability or compensatory mechanisms become overwhelmed. This is consistent with our previous work.12,14
The cohort of 64 preHD gene expansion carriers and 67 controls described here is limited when compared to the larger Track-HD and Predict-HD studies. However, recruiting preHD gene-expansion carriers very far from onset is challenging for a number of reasons. The uptake of genetic testing in this age group is much lower than in those closer to onset83 and this group is much less likely to attend Huntington’s disease clinics regularly, if at all, when compared to preHD gene carriers within 10 years from onset. Tattoos were more common in this age group and both tattoo location and size could result in exclusion from MRI scanning. Finally, this study required participants to agree to undergo lumbar puncture, an invasive procedure.
While there is no clear optimal atlas for connectomics84 we selected the Schaefer cortical resting-state fMRI atlas, which is based on 1489 healthy participants and provides parcellation schemes ranging from 100 to 1000 nodes. We performed NBS and genetic analyses both on the 100 and 500 parcellations to replicate our findings on coarse- and fine-grained atlases. We opted not to use schemes above 500 nodes as connectome reliability decreases considerably, particularly for diffusion MRI-derived structural connectomes, at denser parcellation schemes.73 Both the rsfMRI brain parcellation atlas and the AHBA are derived from the brains of healthy controls. When considering the application of these in our very-far-from-onset preHD gene expansion carrier cohort we must emphasize that detailed multimodal neuroimaging analysis in this cohort has demonstrated that the brain structure is largely normal.19 Furthermore, with respect to regional levels of gene expression and the application of the AHBA atlas, to date, transcriptomic changes in human Huntington’s disease have been demonstrated in post-mortem brains, which are typically at the end stage of the disease, or Huntington’s disease rodent models85 and have upwards of 100 CAG repeats—more representative of the juvenile Huntington’s disease variant.86
This study has characterized functional brain networks in asymptomatic preHD gene carriers very far from disease onset, showing evidence of upregulated functional network connectivity related to disease burden in the presence of normal white matter brain networks. This relationship was found between brain areas that show regional expression of genes specific to neuronal GABAergic and glutamatergic cells following cell-enrichment analysis; a finding that was supported by snRNAseq data from post-mortem Huntington’s disease and healthy control brains that showed an association with neuronal genes abnormally transcribed in Huntington’s disease. In sum, those furthest from Huntington’s disease disease onset display pathology-related functional connectivity changes that are likely characterized by GABAergic inhibition and glutamatergic excitotoxicity.
Supplementary Material
Acknowledgements
We are grateful to the study participants and their families who supported us, and thank the staff at the Wellcome Centre for Human Neuroimaging (London, UK) and the Leonard Wolfson Experimental Neurology Centre (London, UK).
Contributor Information
Peter McColgan, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Sarah Gregory, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Paul Zeun, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Angeliki Zarkali, Dementia Research Centre, University College London, London WC1N 3AR, UK.
Eileanoir B Johnson, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Christopher Parker, Department of Computer Science and Centre for Medical Image Computing, University College London, London WC1V 6LJ, UK.
Kate Fayer, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Jessica Lowe, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Akshay Nair, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; Max Planck University College London Centre for Computational Psychiatry and Ageing Research, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Carlos Estevez-Fraga, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Marina Papoutsi, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Hui Zhang, Dementia Research Centre, University College London, London WC1N 3AR, UK.
Rachael I Scahill, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Sarah J Tabrizi, Huntington’s Disease Centre, Department of Neurodegenerative disease, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; Dementia Research Centre, University College London, London WC1N 3AR, UK.
Geraint Rees, University College London Institute of Cognitive Neuroscience, University College London, London WC1N 3AZ, UK.
Funding
This study was supported by a Wellcome Collaborative Award 200181/Z/15/Z. Funding for CSF collection was provided by the CHDI Foundation, a not-for-profit organization dedicated to finding treatments for Huntington’s disease. S.J.T. is partly supported by the UK Dementia Research Institute that receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK. Some of this work was undertaken at the University College London Hospital/University College London (London, UK) supported by the UK’s Department of Health National Institute of Health Research Biomedical Research Centre (London, UK). We thank Osama Al-Dalahmah and Vilas Menon for providing snRNAseq data.
Competing interests
S.J.T. receives grant funding for her research from the Medical Research Council UK, the Wellcome Trust, the Rosetrees Trust, Takeda Pharmaceuticals Ltd, Vertex Pharmaceuticals, Cantervale Limited, NIHR North Thames Local Clinical Research Network, UK Dementia Research Institute and the CHDI Foundation. Through UCL Consultants Ltd, S.J.T. has undertaken consultancy services for Alnylam Pharmaceuticals Inc., Annexon Inc., DDF Discovery Ltd, F. Hoffmann-La Roche Ltd, Genentech, Novartis Pharma, PTC Therapuetics, Takeda Pharmaceuticals Ltd, Triplet Therapeutics, University College Irvine and Vertex Pharmaceuticals Incorporated. P.M.C. is a full-time employee of F. Hoffmann-La Roche.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Tabrizi SJ, Reilmann R, Roos RA, et al. . Potential endpoints for clinical trials in premanifest and early Huntington’s disease in the TRACK-HD study: Analysis of 24 month observational data. Lancet Neurol. 2012;11:42–53. [DOI] [PubMed] [Google Scholar]
- 2. Gregory S, Crawford H, Seunarine K, et al. . Natural biological variation of white matter microstructure is accentuated in Huntington’s disease. Hum Brain Mapp. 2018;39:3516–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Georgiou-Karistianis N, Scahill R, Tabrizi SJ, Squitieri F, Aylward E. Structural MRI in Huntington’s disease and recommendations for its potential use in clinical trials. Neurosci Biobehav Rev. 2013;37:480–490. [DOI] [PubMed] [Google Scholar]
- 4. Johnson EB, Gregory S. Huntington’s disease: Brain imaging in Huntington’s disease. Prog Mol Biol Transl Sci. 2019;165:321–369. [DOI] [PubMed] [Google Scholar]
- 5. Poudel GR, Stout JC, Domínguez DJ, et al. . Functional changes during working memory in Huntington’s disease: 30-month longitudinal data from the IMAGE-HD study. Brain Struct Funct. 2015;220:501–512. [DOI] [PubMed] [Google Scholar]
- 6. Werner CJ, Dogan I, Saß C, et al. . Altered resting-state connectivity in Huntington’s disease. Hum Brain Mapp. 2014;35:2582–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolf RC, Sambataro F, Vasic N, et al. . Longitudinal task-negative network analyses in preclinical Huntington’s disease. Eur Arch Psychiatry Clin Neurosci. 2014;264:493–505. [DOI] [PubMed] [Google Scholar]
- 8. Wolf RC, Sambataro F, Vasic N, et al. . Default-mode network changes in preclinical Huntington’s disease. Exp Neurol. 2012;237:191–198. [DOI] [PubMed] [Google Scholar]
- 9. Gargouri F, Messé A, Perlbarg V, et al. . Longitudinal changes in functional connectivity of cortico-basal ganglia networks in manifests and premanifest Huntington’s disease. Hum Brain Mapp. 2016;37:4112–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Georgiou-Karistianis N, Poudel GR, Domínguez DJ, et al. . Functional and connectivity changes during working memory in Huntington’s disease: 18 month longitudinal data from the IMAGE-HD study. Brain Cogn. 2013;83:80–91. [DOI] [PubMed] [Google Scholar]
- 11. Langley C, Gregory S, Osborne-Crowley K, et al. . Fronto-striatal circuits for cognitive flexibility in far from onset Huntington’s disease: Evidence from the Young Adult Study. J Neurol Neurosurg Psychiatry. 2021;92:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McColgan P, Gregory S, Razi A, et al. . White matter predicts functional connectivity in premanifest Huntington’s disease. Ann Clin Transl Neurol. 2017;4(2):106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gregory S, Long JD, Klöppel S, et al. . Operationalizing compensation over time in neurodegenerative disease. Brain. 2017;140(4):1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gregory S, Long JD, Klöppel S, et al. . Testing a longitudinal compensation model in premanifest Huntington’s disease. Brain. 2018;141(7):2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klöppel S, Gregory S, Scheller E, et al. . Compensation in preclinical Huntington’s disease: Evidence from the track-on HD study. EBioMedicine. 2015;2:1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Gregory S, Scahill RI, et al. . In vivo characterization of white matter pathology in pre-manifest Huntington’s disease. Ann Neurol. 2018;84:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McColgan P, Seunarine KK, Razi A, et al. . Selective vulnerability of Rich Club brain regions is an organizational principle of structural connectivity loss in Huntington’s disease. Brain. 2015;138(11):3327–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McColgan P, Gregory S, Seunarine KK, et al. . Brain regions showing white matter loss in Huntington’s disease are enriched for synaptic and metabolic genes. Biol Psychiatry. 2018;83:456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scahill RI, Zeun P, Osborne-Crowley K, et al. . Biological and clinical characteristics of gene carriers far from predicted onset in the Huntington’s disease Young Adult Study (HD-YAS): A cross-sectional analysis. Lancet Neurol. 2020;19(6):502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeun P, McColgan P, Dhollander T, et al. Timing of selective basal ganglia white matter loss in Huntington’s disease. bioRxiv. [Preprint] doi: 10.1101/2021.02.17.431568 [DOI] [PMC free article] [PubMed]
- 21. Šišková Z, Justus D, Kaneko H, et al. . Dendritic structural degeneration is functionally linked to cellular hyperexcitability in a mouse model of Alzheimer’s disease. Neuron. 2014;84:1023–1033. [DOI] [PubMed] [Google Scholar]
- 22. Callahan JW, Abercrombie ED. Age-dependent alterations in the cortical entrainment of subthalamic nucleus neurons in the YAC128 mouse model of Huntington’s disease. Neurobiol Dis. 2015;78:88–99. [DOI] [PubMed] [Google Scholar]
- 23. Ravalia AS, Lau J, Barron J, et al. . Super-resolution imaging reveals extrastriatal synaptic dysfunction in presymptomatic Huntington disease mice. Neurobiol Dis. 2021;152:105293. [DOI] [PubMed] [Google Scholar]
- 24. Vucic S, Nicholson GA, Kiernan MC. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain. 2008;131:1540–1550. [DOI] [PubMed] [Google Scholar]
- 25. Cheng A, Wang J, Ghena N, et al. . SIRT3 haploinsufficiency aggravates loss of GABAergic interneurons and neuronal network hyperexcitability in an Alzheimer’s disease model. J Neurosci. 2020;40:694–709 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu D, Yuan T, Gao J, et al. . Correlation between cortical gene expression and resting-state functional network centrality in healthy young adults. Hum Brain Mapp. 2021;42:2236–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chamizo VD, Rodríguez CA, Sánchez J, Mármol F. Sex differences after environmental enrichment and physical exercise in rats when solving a navigation task. Learn Behav. 2016;44:227–238. [DOI] [PubMed] [Google Scholar]
- 28. Xue K, Liang S, Yang B, et al. . Local dynamic spontaneous brain activity changes in first-episode, treatment-naïve patients with major depressive disorder and their associated gene expression profiles. Psychol Med. 2020:1–10. doi: 10.1017/S0033291720003876 [DOI] [PubMed] [Google Scholar]
- 29. Zarkali A, McColgan P, Ryten M, et al. . Dementia risk in Parkinson’s disease is associated with interhemispheric connectivity loss and determined by regional gene expression. NeuroImage Clin. 2020;28:102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seredenina T, Luthi-Carter R. What have we learned from gene expression profiles in Huntington’s disease? Neurobiol Dis. 2012;45:83–98. [DOI] [PubMed] [Google Scholar]
- 31. Hodges A, Strand AD, Aragaki AK, et al. . Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. [DOI] [PubMed] [Google Scholar]
- 32. Byrne LM, Rodrigues FB, Johnson EB, et al. . Evaluation of mutant huntingtin and neurofilament proteins as potential markers in Huntington’s disease. Sci Transl Med. 2018;10(458):eaat7108. [DOI] [PubMed] [Google Scholar]
- 33. Johnson EB, Byrne LM, Gregory S, et al. . Neurofilament light protein in blood predicts regional atrophy in Huntington disease. Neurology. 2018;90:e717–e723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Byrne LM, Rodrigues FB, Blennow K, et al. . Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington’s disease: A retrospective cohort analysis. Lancet Neurol. 2017;16:601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR, International Huntington’s Disease Collaborative Group . A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65:267–277. [DOI] [PubMed] [Google Scholar]
- 36. Unified Huntington’s disease rating scale: Reliability and consistency. Huntington Study Group. Mov Disord. 1996;11(2):136–142. [DOI] [PubMed] [Google Scholar]
- 37. Wild EJ, Boggio R, Langbehn D, et al. . Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J Clin Invest. 2015;125:1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaefer A, Kong R, Gordon EM, et al. . Local–global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28:3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi EY, Thomas Yeo BT, Buckner RL. The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol. 2012;108:2242–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith RE, Tournier JD, Calamante F, Connelly A. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage. 2012;62(3):1924–1938. [DOI] [PubMed] [Google Scholar]
- 41. Tournier J-D, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol. 2012;22(1):53–66. [Google Scholar]
- 42. Veraart J, Novikov DS, Christiaens D, Ades-aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage. 2016;142:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med. 2016;76(5):1574–1581. [DOI] [PubMed] [Google Scholar]
- 44. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tustison NJ, Avants BB, Cook PA, et al. . N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage. 2014;103:411–426. [DOI] [PubMed] [Google Scholar]
- 47. Smith RE, Tournier JD, Calamante F, Connelly A. SIFT: Spherical-deconvolution informed filtering of tractograms. Neuroimage. 2013;67:298–312. [DOI] [PubMed] [Google Scholar]
- 48. Smith RE, Tournier JD, Calamante F, Connelly A. The effects of SIFT on the reproducibility and biological accuracy of the structural connectome. Neuroimage. 2015;104:253–265. [DOI] [PubMed] [Google Scholar]
- 49. Whitfield-Gabrieli S, Nieto-Castanon A. Conn: A functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. [DOI] [PubMed] [Google Scholar]
- 50. Lindquist MA, Geuter S, Wager TD, Caffo BS. Modular preprocessing pipelines can reintroduce artifacts into fMRI data. Hum Brain Mapp. 2019;40(8):2358–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hallquist MN, Hwang K, Luna B. The nuisance of nuisance regression: Spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage. 2013;82:208–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Polosecki P, Castro E, Rish I, et al. . Resting-state connectivity stratifies premanifest Huntington’s disease by longitudinal cognitive decline rate. Sci Rep. 2020;10(1):1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zalesky A, Fornito A, Bullmore ET. Network-based statistic: Identifying differences in brain networks. Neuroimage. 2010;53:1197–1207. [DOI] [PubMed] [Google Scholar]
- 54. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. . An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arnatkevičiūtė A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. bioRxiv. [Preprint]. doi:10.1101/380089
- 56. Romero-Garcia R, Whitaker KJ, Váša F, et al. . Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage. 2018;171:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Markello RD, Arnatkeviciute A, Poline J-B, Fulcher BD, Fornito A, Misic B. Standardizing workflows in imaging transcriptomics with the abagen toolbox. Elife. 2021;10:e72129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin J, French L, Xu T, et al. . Cell-specific gene-expression profiles and cortical thickness in the human brain. Cereb Cortex. 2018;28(9):3267–3277. [DOI] [PubMed] [Google Scholar]
- 59. Cao H, Plichta MM, Schäfer A, et al. . Test–retest reliability of fMRI-based graph theoretical properties during working memory, emotion processing, and resting state. Neuroimage. 2014;84:888–900. [DOI] [PubMed] [Google Scholar]
- 60. Patel Y, Shin J, Gowland PA, Pausova Z, Paus T. Maturation of the human cerebral cortex during adolescence: Myelin or dendritic arbor? Cereb Cortex. 2019;29(8):3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Alexander-Bloch A, Shou H, Liu S, et al. . On testing for spatial correspondence between maps of human brainstructure and function. Neuroimage. 2018;178:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas GEC, Zarkali A, Ryten M, et al. . Regional brain iron and gene expression provide insights into neurodegeneration in Parkinson’s disease. Brain. 2021;144(6):1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Whitaker KJ, Vértes PE, Romero-Garciaa R, et al. . Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci USA. 2016;113:9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vértes PE, Rittman T, Whitaker KJ, et al. . Gene transcription profiles associated with inter-modular hubs and connection distance in human functional magnetic resonance imaging networks. Philos Trans R Soc B Biol Sci. 2016;371:20150362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Skene NG, Grant SGN. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front Neurosci. 2016;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jia P, Tian J, Zhao Z. Assessing gene length biases in gene set analysis of genome-wide association studies. Int J Comput Biol Drug Des. 2010;3(4):297–310. [DOI] [PubMed] [Google Scholar]
- 67. Hawrylycz M, Miller JA, Menon V, et al. . Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Habib N, Avraham-Davidi I, Basu A, et al. . Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Langfelder P, Cantle JP, Chatzopoulou D, et al. . Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Durrenberger PF, Fernando FS, Kashefi SN, et al. . Common mechanisms in neurodegeneration and neuroinflammation: A BrainNet Europe gene expression microarray study. J Neural Transm. 2015;122:1055–1068. [DOI] [PubMed] [Google Scholar]
- 71. Zhang B, Gaiteri C, Bodea LG, et al. . Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Al-Dalahmah O, Sosunov AA, Shaik A, et al. . Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol Commun. 2020;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yeh CH, Smith RE, Hollander T, Calamante F, Connelly A.. Investigating the mechanisms of structural connectome construction on HCP data: Implications for structural network analysis. ISMRM. https://www.researchgate.net/publication/307862957_Investigating_the_mechanisms_of_structural_connectome_construction_on_HCP_data_implications_for_structural_network_analysis. Accessed 12 April 2021.
- 74. Papoutsi M, Labuschagne I, Tabrizi SJ, Stout JC. The cognitive burden in Huntington’s disease: Pathology, phenotype, and mechanisms of compensation. Mov Disord. 2014;29:673–683. [DOI] [PubMed] [Google Scholar]
- 75. Stout JC, Jones R, Labuschagne I, et al. . Evaluation of longitudinal 12 and 24 mo7nth cognitive outcomes in premanifest and early Huntington’s disease. J Neurol Neurosurg Psychiatry. 2012;83:687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Behrens PF, Franz P, Woodman B, Lindenberg KS, Landwehrmeyer GB. Impaired glutamate transport and glutamate–glutamine cycling: Downstream effects of the Huntington mutation. Brain. 2002;125:1908–1922. [DOI] [PubMed] [Google Scholar]
- 77. Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133:257–261. [DOI] [PubMed] [Google Scholar]
- 78. Spampanato J, Gu X, Yang XW, Mody I. Progressive synaptic pathology of motor cortical neurons in a BAC transgenic mouse model of Huntington’s disease. Neuroscience. 2008;157:606–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim EH, Thu DCV, Tippett LJ, et al. . Cortical interneuron loss and symptom heterogeneity in Huntington disease. Ann Neurol. 2014;75:717–727. [DOI] [PubMed] [Google Scholar]
- 80. Mehrabi NF, Waldvogel HJ, Tippett LJ, Hogg VM, Synek BJ, Faull RLM. Symptom heterogeneity in Huntington’s disease correlates with neuronal degeneration in the cerebral cortex. Neurobiol Dis. 2016;96:67–74. [DOI] [PubMed] [Google Scholar]
- 81. Thu DCV, Oorschot DE, Tippett LJ, et al. . Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington’s disease. Brain. 2010;133:1094–1110. [DOI] [PubMed] [Google Scholar]
- 82. Macdonald V, Halliday G. Pyramidal cell loss in motor cortices in Huntington’s disease. Neurobiol Dis. 2002;10:378–386. [DOI] [PubMed] [Google Scholar]
- 83. Baig SS, Strong M, Rosser E, et al. . 22 Years of predictive testing for Huntington’s disease: The experience of the UK Huntington’s Prediction Consortium. Eur J Hum Genet. 2016;24:1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Arslan S, Ktena SI, Makropoulos A, Robinson EC, Rueckert D, Parisot S. Human brain mapping: A systematic comparison of parcellation methods for the human cerebral cortex. Neuroimage. 2018;170:5–30. [DOI] [PubMed] [Google Scholar]
- 85. Bragg RM, Coffey SR, Weston RM, et al. . Motivational, proteostatic and transcriptional deficits precede synapse loss, gliosis and neurodegeneration in the B6.HttQ111/+ model of Huntington’s disease. Sci Rep. 2017;7:41570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat Rev Neurosci. 2013;14:708–721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized, derived data supporting the findings of this study are available from the corresponding author on request. Code used to implement analyses in this study is freely available at https://github.com/AngelikaZa/YAS_HD.