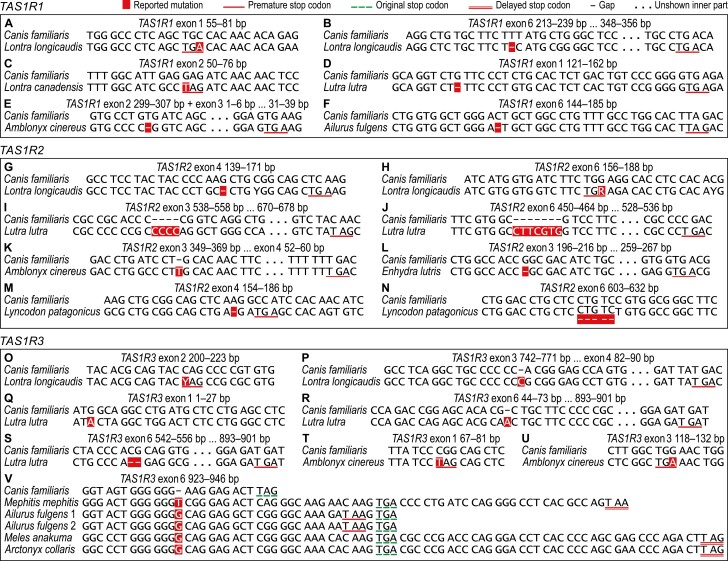
Fig. 1.
DNA sequence alignments showing the start codon, nonsense, and frameshift mutations found in this study. (A) Homozygous substitution of C to A at 69 bp of TAS1R1 exon 1 in Lontra longicaudis resulting in a premature homozygous stop codon (67–69 bp). (B) 1-bp deletion at 226 bp of TAS1R1 exon 6 in Lontra longicaudis resulting in multiple premature homozygous stop codons with the first one at 352–354 bp. (C) Homozygous substitution of G to T at 62 bp of TAS1R1 exon 2 in Lontra canadensis resulting in a premature homozygous stop codon (62–64 bp). (D) 1-bp deletion at 129 bp of TAS1R1 exon 1 in Lutra lutra resulting in multiple premature homozygous stop codons with the first one at 158–160 bp of exon 1. (E) 1-bp deletion at 306 bp of TAS1R1 exon 2 in Amblonyx cinereus resulting in multiple premature homozygous stop codons with the first one at 35–37 bp of exon 3. (F) 1-bp deletion at 157 bp of TAS1R1 exon 6 in Ailurus fulgens resulting in multiple premature homozygous stop codons with the first one at 181–183 bp. (G) 1-bp deletion at 156 bp of TAS1R2 exon 4 in Lontra longicaudis resulting in multiple premature homozygous stop codons with the first one at 167–169 bp of exon 4. (H) Heterozygous substitution of G to A at 173 bp of TAS1R2 exon 6 in Lontra longicaudis resulting in a premature heterozygous stop codon (TGA at 171–173 bp). (I) 4-bp insertion between 547 and 548 bp of TAS1R2 exon 3 in Lutra lutra resulting in multiple premature homozygous stop codons with the first one at 675–677 bp of exon 3. (J) 7-bp insertion between 457 and 458 bp of TAS1R2 exon 6 in Lutra lutra resulting in multiple premature homozygous stop codons with the first one at 533–535 bp. (K) 1-bp insertion between 359 and 360 bp of TAS1R2 exon 3 in Amblonyx cinereus (LC654484 and JN130352) resulting in multiple premature homozygous stop codons with the first one at 57–59 bp of exon 4. (L) 1-bp deletion at 205 bp of TAS1R2 exon 3 in Enhydra lutris resulting in multiple premature homozygous stop codons with the first one at 263–265 bp of exon 3. (M) 1-bp deletion at 170 bp of TAS1R2 exon 4 in Lyncodon patagonicus resulting in multiple premature homozygous stop codons with the first one at 173–175 bp of exon 4. (N) 5-base deletion at 615–619 bp of TAS1R2 exon 6 in Lyncodon patagonicus not resulting in a premature stop codon. (O) Heterozygous substitution of C to T at 212 bp of TAS1R3 exon 2 in Lontra longicaudis resulting in a premature heterozygous stop codon (TAG at 212–214 bp). (P) 1-bp insertion between 758 and 759 bp of TAS1R3 exon 3 in Lontra longicaudis resulting in multiple premature homozygous stop codons with the first one at 87–89 bp of exon 4. (Q) Homozygous substitution of G to A in the start codon of TAS1R3 in Lutra lutra (nearest downstream in-frame ATG sequence is at 85–87 bp of exon 1). (R) 1-bp insertion between 60 and 61 bp of TAS1R3 exon 6 in Lutra lutra resulting in a premature homozygous stop codon at 898–900 bp. (S) 2-bp deletion at 549 and 550 bp of TAS1R3 exon 6 in Lutra lutra resulting in a premature homozygous stop codon at 898–900 bp. (T) Homozygous substitution of C to T at 73 bp of TAS1R3 exon 1 in Amblonyx cinereus resulting in a premature homozygous stop codon (73–75 bp). (U) Homozygous substitution of G to A at 126 bp of TAS1R3 exon 3 in Amblonyx cinereus resulting in a premature homozygous stop codon (124–126 bp). (V) 1-bp insertion between 934 and 935 bp of TAS1R3 exon 6 in Mephitis mephitis, Ailurus fulgens (1, LC654495; 2, LNAC01000817), Meles anakuma, and Arctonyx collaris resulting in a premature (Ailurus fulgens) or delayed (remaining species) homozygous stop codon; note that the arctoid TAS1R3 exon 6 is 5 codons longer than the canid one (this study; Wolsan and Sato 2020). Base-pair position numbering refers to the aligned Canis familiaris sequence, starts from the 5ʹ end of each exon separately, and is from left to right. Codons in the correct open reading frame are separated by spaces.