Highlights
-
•
Established fluorescent probe-based RPA (exo RPA) and RPA-LFD methods for fast and sensitive detection of E. miricola.
-
•
Exo RPA and RPA-LFD could detect E. miricola genomic DNA at 38 °C in 30 min.
-
•
The detection sensitivity of exo RPA and RPA-LFD were 102 copies/μL.
-
•
The exo RPA and RPA-LFD achieved rapid detection of E. miricola without bulky lab equipment.
Keywords: Elizabethkingia miricola, Recombinase polymerase amplification (RPA), Lateral flow dipstick, exo RPA, Pelophylax nigromaculatus
Abstract
Elizabethkingia miricola is a highly infectious pathogen, which causes high mortality rate in frog farming. Therefore, it is urgent to develop a rapid and sensitive detection method. In this study, two rapid and specific methods including recombinase polymerase amplification combined with lateral flow dipstick (RPA-LFD) and fluorescent probe-based recombinase polymerase amplification (exo RPA) were established to effectively detect E. miricola, which can accomplish the examination at 38 °C within 30 min. The limiting sensitivity of RPA-LFD and exo RPA (102 copies/μL) was ten-fold higher than that in generic PCR assay. The specificities of the two methods were verified by detecting multiple DNA samples (E. miricola, Staphylococcus aureus, Aeromonas hydrophila, Aeromonas veronii, CyHV-2 and Edwardsiella ictaluri), and the result showed that the single band was displayed in E. miricola DNA only. By tissue bacterial load and qRT-PCR assays, brain is the most sensitive tissue. Random 24 black spotted frog brain samples from farms were tested by generic PCR, basic RPA, RPA-LFD and exo RPA assays, and the results showed that RPA-LFD and exo RPA methods were able to detect E. miricola accurately and rapidly. In summary, the methods of RPA-LFD and exo RPA were able to detect E. miricola conveniently, rapidly, accurately and sensitively. This study provides prospective methods to detect E. miricola infection in frog culture.
1. Introduction
Elizabethkingia miricola is a gram-negative bacterium that can cause a variety of diseases in humans and animals [1]. E. miricola causes joint infections in multiple anuran (frogs and toads) species [2]. In addition, According to previous studies, E. miricola was proposed to cause urinary tract and native joint infections in humans [3,4]. Pelophylax nigromaculatus (black spotted frog) culture is booming because of delicious taste, abundant nutrition and high market value [5]. However, intensive aquaculture brings about a parallel growth trend of a variety of infectious diseases [6]. Black spotted frogs with E. miricola infection show the symptom of distorted head, cataracts and red lips [7]. The optimal time to control the diseases was easily missed, resulting in huge economic losses in the process of aquaculture. Therefore, it is essential to develop a rapid and convenient diagnostic method to detect potential E. miricola for the frog farming industry.
In general, traditional PCR has shown some advantages in rapid detection of bacteria [8,9]. However, traditional PCR keeps some defects. The traditional PCR is time-consuming and requires expensive temperature-controlled equipment. The real-time PCR and loop-mediated isothermal amplification (LAMP) have been further developed for detection of bacteria [10], [11], [12]. However, the reactions are affected by polymerase inhibitors possibly, and require temperature-controlled equipment and skilled operators. Therefore, there is an urgent need for convenient and rapid methods for clinical detection.
Recombinase polymerase amplification (RPA) technology, first introduced in 2006, is a novel isothermal nucleic acid amplification technique, which mainly relies on three proteins: recombinase that binds single-stranded nucleic acids (oligonucleotide primers), single-stranded DNA-binding protein (SSB), and strand-substituting DNA polymerase [13]. RPA, a simple, rapid, and low cost without the need of sophisticated equipment or specialized skill, is of great application. For instance, RPA is used to detect genotype III grass carp reovirus [14], shrimp hemocyte iridescent virus [15] and spring viremia of carp virus [16]. Previous reports described that RPA was more sensitive, rapid and accurate than traditional PCR [17]. Although the traditional RPA can achieve the detection of RPA products by agarose gel electrophoresis, the operation process is prone to cross-contamination and is not suitable for clinical detection [18].
Recombinase polymerase amplification combined with lateral flow dipstick (RPA-LFD) is an innovative detection method suitable for practical application. With the advantages of visual and handy, RPA-LFD has received extensive attention [17,19,20]. This method not only avoids the possibility of cross-contamination, but also eliminates the need for bulky laboratory equipment and cumbersome handling steps. The results are visualized in less than 30 min. RPA-LFD is used to detect infectious spleen and kidney necrosis virus [17], cyprinid herpesvirus 2 [20], African swine fever virus [19], goatpox virus and sheeppox virus [21], Escherichia coli O157:H7 [22] and salmonella spp. in food samples [23].
Fluorescent probe-based recombinase polymerase amplification (exo RPA) is the basic RPA combined with the exo probe technology [24], which can be imaged with a FAM channel to observe the presence or absence of fluorescence to determine whether the target gene is detected. The portable machine for detecting the fluorescence of exo RPA products is inexpensive and practical, which is suitable for field detection. Exo RPA is used to detect cucumber mosaic virus in banana plants [24], Orf virus [25] and Salmonella enterica [26], Haemophilus parasuis [27], shrimp white spot syndrome virus [28] and E. coli [29].
However, the detection of E. miricola by RPA has not been reported. In the present study, we aimed to develop two rapid and sensitive methods (RPA-LFD and exo RPA) for the detection of E. miricola. Screening suitable primers and probes were crucial in RPA-LFD and exo RPA. According to previous study, the mutT was specific and conserved gene in E. miricola through comparison and screening of a number of genes [12]. In this study, we selected the mutT gene sequence of E. miricola as the target sequence (GenBank accession number: NZ-CP040516.1). The primers and probes for detection of E. miricola were designed based on target sequence. RPA-LFD and exo RPA are promising and provide rapid and sensitive detection of pathogenic bacteria. The analytical sensitivity and specificity were evaluated. The applicability was determined by detecting clinical samples.
2. Materials and methods
2.1. Frogs, pathogens and sampling
200 black spotted frogs were obtained from farms with and without infection history of wryneck disease in Wuhan, Hubei Province. The frogs were evenly divided into five tanks (83 cm × 60 cm × 54 cm). The tanks were kept wet. All the experimental procedures were approved by the animal protection and used Committee of Huazhong Agricultural University. The ethical number is HZAUFR-2021–0001.
E. miricola, Aeromonas hydrophila (ATCC 7966), Staphylococcus aureus (ATCC 25,923), Aeromonas veronii (LMG 9075), Edwardsiella ictalurid (DSM 13,697) and CyHV-2 were from our laboratory stock [30], [31], [32]. E. miricola strain was isolated and identified by our laboratory. The E. miricola strain was cultured to mid-log stage in brain–heart infusion (BHI) medium (Haibo biology, Oxoid Ltd) for 24 h at 37 °C. Other pathogens were cultured to mid-log stage in Luria-Bertani (LB, L3027, Sigma, Shanghai, China) medium. CyHV-2 was kept at −80 °C.
Black spotted frogs with significant distorted head and cataracts were selected for the tissue bacterial load and qRT-PCR assays. Tissues from healthy black spotted frogs were used as negative control. The frogs were enesthetized with 0.4 g/L MS-222 (3-Aminobenzoic acid ethyl ester methanesulfonate) (Merck, Germany) for 20 min and their tissues (liver, lung, brain, stomach, spleen, heart, kidney, eye, and muscle) were dissected in a sterile environment.
2.2. DNA extraction
Tissue DNA was extracted from dissecting black spotted frogs. The samples were cut into mung bean size with scissors, and then grinded 5 min with 150 μL DNA extraction buffer (0.5% sodium dodecyl sulfate,1 μg/mL, 300 mM NaCl 6 mL, 10 mM Tris–HCl, 2 mL, pH 8.0, 10 mM EDTA, 10 mL, pH 8.0). Then centrifuged slightly and added 10 μL proteinase K (150 μg/mL), and the samples were digested in 62 °C for 3 h. Then 600 μL Tris-saturated phenol was added and fully mixed for 10 min, and centrifugation was performed at 12,000 rpm for 10 min. The supernatant was transferred to a new EP tube, twice the volume of ethanol was added, precipitated at −20 °C for 30 min, and centrifuged at 12,000 rpm for 5 min. The supernatant was discarded, then the precipitate was washed twice with 70% ethanol and air-dried at room temperature. 50 μL the extracted DNA was dissolved in ultrapure water and then was stored at −20 °C for standby.
2.3. Designing primers and probes
The mutT gene of E. miricola was selected as target sequence. We designed three forward primers and three reverse primers according to the principles of RPA primer (generally the length of RPA primer is 30–35 nt, the amplification efficiency is high, and the amplification product is less than 500 bp) and examined nine primer combinations (mutTF699/mutTR700, mutTF699/mutTR702, mutTF699/mutTR704 mutTF701/mutTR702, mutTF701/ mutTR700, mutTF701/ mutTR704, mutTF703/mutTR704, mutTF703/mutTR700, mutTF703/ mutTR702). The results of RPA reaction were detected with 2% agarose gel.
The RPA-LFD probe was designed according to the sequence between the optimal primers. The RPA-LFD probe added a FAM on the 5′ end, a C3 spacer on the 3′ end, a dSpacer (tetrahydrofuran, THF) in the middle, and the reverse primer for RPA-LFD added a biotin on the 5′ end.
The probe for exo RPA was designed based on the sequence between the optimal primers. The exo RPA probe THF as the recognition site of exonuclease is located at least 35 nt away from the 5′ end. Then, a fluorescent group was labeled at the upstream of THF site, and a quenched group was labeled at the downstream. The distance between the two groups is 1–4 nt [33]. All primers and probes in this study were listed in Table 1, were synthesized by Tsingke Biotechnology (Beijing, China).
Table 1.
Primers and probes in this study.
| Method | Primername | Primerdirection | Sequence (5′–3′) | Location (CP040516.1) | |
|---|---|---|---|---|---|
| RPA | mutTF699 | Forward | AAGAGGAACTCGGAATAAGGGTTGAAATAG | 132–161 | |
| mutTR700 | Reverse | ATACCTTTCTATTTCAACCCTTATTCCGAGT | 173–198 | ||
| mutTF701 | Forward | CCTGAAAAATCATTGGGTGGTTATTGGGAG | 41–70 | ||
| mutTR702 | Reverse | CCATAGAACACAAAACATCAGCAATAGGAAT | 355–385 | ||
| mutTF703 | Forward | ATGGGTACGATTAGGGTTGTTTGTGGTGTT | 1–30 | ||
| mutTR704 | Reverse | ATGATCGGTGAGTACGCAAATGGACTGTTC | 265–294 | ||
| PCR | mutTF705 | Forward | TTGCCTCTGCCCGAATC | 172,228–172,244 | |
| mutTR706 | Reverse | TTACACATCCATAGAACACAAAACA | 172,766–172,790 | ||
| RPA-LFD | mutTF703 | Forward | ATGGGTACGATTAGGGTTGTTTGTGGTGTT | 1–30 | |
| mutTR700 | Reverse | (Biotin)ACTCGGAATAAGGGTTGAAATAGAAAGGTAT | 173–198 | ||
| mutT1 | probe | (FAM)TATTCAATGAAGGACGAATTTTGCTTTGTAG-THF-CGTAAGCCTGAAAAATCA (C3spacer) | |||
| exo RPA | mutTF703 | Forward | ATGGGTACGATTAGGGTTGTTTGTGGTGTT | 1–30 | |
| mutTR700 | Reverse | ACTCGGAATAAGGGTTGAAATAGAAAGGTAT | 173–198 | ||
| mutT2 | probe | ATATTCAATGAAGGACGAATTTTGCTTTG (FAM-T)AG-THF-CG(BHQ1-T)AAGCCTGAAAAATCA (C3spacer) | |||
| qRT-PCR | mutTF707 | Forward | CGTATATATGTAGGTCGGAACAG | [12] | |
| mutTF708 | Reverse | CCATAGAACACAA AACATCAGCA | |||
| 18SF709 | Forward | CGTTGATTAAGTCCCTGCCCTT | [7] | ||
| 18SR710 | Reverse | GCCGATCCGAGGACCTCACTA | |||
2.4. Establishment of basic RPA, RPA-LFD and exo RPA assays
The basic RPA was initiated by using the TwistAmp Basic kit (TwistDx, Cambridge, UK), and the reaction system contained 29.5 μL rehydration buffer, 2 μL forward primer, 2 μL reverse primer, 12 μL ddH2O and 2 μL E. miricola DNA sample, and then 2.5 μL magnesium acetate was added immediately to start the reaction, and the reaction lasted for 30 min at 38 °C [17]. H2O was used as a negative control. The reaction products were purified by phenol chloroform 1:1 extract and analyzed on 2% agarose gel electrophoresis subsequently.
TwistAmp nfo kit (TwistDX, Cambridge, UK) and Hybridetect 1 (Milenia Biotec GmbH, GieBen, Germany) dipsticks were used for RPA-LFD. The reaction system included 29.4 μL rehydration buffer, 2 μL forward primer, 2 μL reverse primer, 0.6 μL probe, 11.5 μL ddH2O and 2 μL E. miricola DNA sample and was started by 2.5 μL magnesium acetate, H2O was used as a negative control. mutT1 probe (without DNA sample) was used as a blank control. RPA-LFD reactions were performed in 38 °C for 30 min [17], and then, the products were diluted at 1:90 with ddH2O. Dipsticks were put into the diluted samples and the results can be read in 5 min.
The exo RPA was initiated by using the TwistAmp exo kit (TwistDx, Cambridge, UK) and the reaction system included 29.4 μL rehydration buffer, 2 μL forward primer, 2 μL reverse primer, 0.6 μL probe, 11.5 μL ddH2O and 2 μL E. miricola DNA sample and was started by 2.5 μL magnesium acetate, H2O was used as a negative control. Fluorescent reactions were performed at 38 °C for 30 min [17]. The fluorescence be observed by fluorescence imaging system (iQ5, Bio-Rad, California, USA) with fluorescence measurements recorded in the FAM channel.
2.5. Specificities of basic RPA, RPA-LFD and exo RPA assays
The genomic DNA were extracted from the pathogens (E. miricola, S. aureus, A. hydrophila, A. veronii, CyHV-2 and E. ictaluri) by a Universal Genomic DNA kit (CWBIO, China). The specificities of basic RPA, RPA-LFD and exo RPA were verified by detecting multiple DNA samples and a negative control (H2O). The fluorescence value of exo RPA product was measured by the enzyme-labeled instrument (Tecan Nano Quant) (wavelength range: 485–535 nm).
2.6. Sensitivities of basic RPA, RPA-LFD and exo RPA assays
The partial sequence (545 bp) of mutT gene of E. miricola was obtained through PCR and cloned into the pMD19-T plasmid to generate recombinant plasmid pMD19-mutT. Then the plasmid was purified and converted to copy number by measuring the concentration of recombinant plasmid, DNA copies/μL = (ng/μL × 6.02 × 1023 × 10−9)/(Fragment length (bp) × 660) [21]. pMD19-mutT recombinant plasmid was diluted from 10° to 107 copies/μL. Detection results of PCR, RPA, RPA-LFD and exo RPA were compared, and the sensitivities of the four methods were analyzed.
2.7. Tissue bacterial load and qRT-PCR of E. miricola
Tissues (liver, lung, brain, stomach, spleen, heart, kidney, eye, and muscle) from five diseased black spotted frogs (distorted head or cataracts) were weighed and then homogenized in a sterile environment. Tissues from healthy black spotted frogs were used as negative control. Per gram of tissues were immediately homogenized in 6 mL PBS buffer by a tissue grinder (P0485, Sigma, Shanghai, China) at 28 °C for 5 min, and then incubated in BHI medium at 28 °C for 24 h. The colonies of E. miricola were pale yellow in color, slightly opaque in BHI medium [6]. The number of bacteria in the tissues was counted by two independent investigators.
Tissue bacterial load was calculated by dilution coated plate method [31]. Frogs euthanized by MS-222 was dissected, and the tissues were immediately gathered and placed into EP tubes containing 800 μL of Trizol. Total RNA was extracted and reversed to cDNA and stored at −80 °C. The bacterial gene expression was analyzed by qRT-PCR, which was performed in a real-time PCR system (Applied Biosystems, USA). The qRT-PCR mixture contained 4 μL of cDNA sample, 3.1 μL of nuclease-free water, 7.5 μL of 2 × AceQ® qPCR SYBR Green Master Mix, and 0.2 μL of each gene specific primer. Conditions for amplification were 3 min at 95 °C, followed by 40 cycles of 15 s at 95 °C (denaturation), 15 s at 60 °C (annealing) and 20 s at 72 °C (extension). 18S rRNA was used as the housekeeping gene. and the relative mRNA expression level was calculated with the 2−ΔΔCT method. All the experiments were performed in triplicate.
2.8. Practical evaluation of RPA-LFD and exo RPA assays
DNA extraction from 24 black spotted frogs with or without E. miricola were randomly selected from frog farms for testing the practical performance of PCR, RPA, RPA-LFD and exo RPA, and DNA was prepared as previously described.
2.9. Statistical analysis
The results were expressed as the means ± standard deviation (SD) and all statistical analysis were done using SPSS 26.0 package. The experimental data were subjected by Dunn's multiple comparison (with Bonferroni adjustment) to identify the significance.
3. Results
3.1. Screening primers and probes
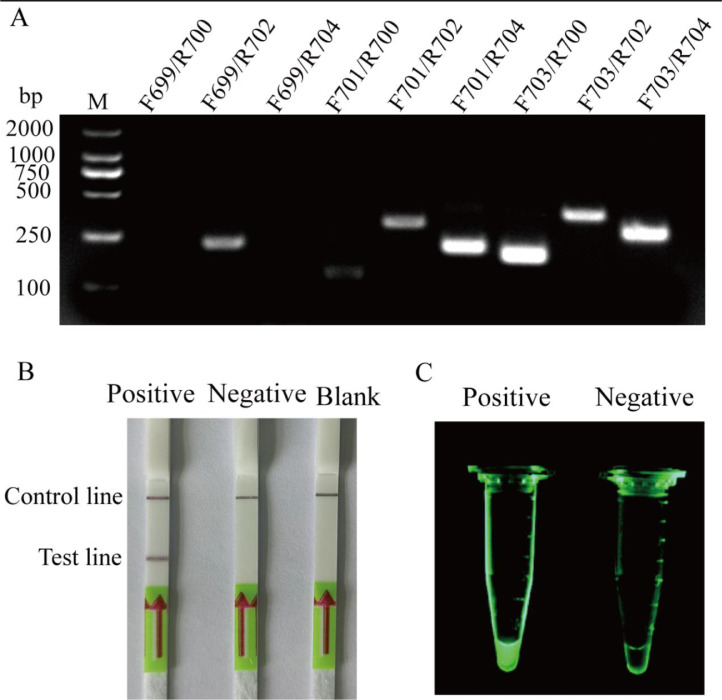
The mutT sequence was selected for designing primers and probes. In the primer screening assay, we designed three forward primers and three reverse primers, and performed nine combinations for RPA test. The agarose gel electrophoresis results showed that the band from mutTF703/mutTR700 primer pair combination was the brightest and single band (Fig. 1A). Therefore, primer pair mutTF703/mutTR700 was optimal for RPA assay. Target product amplified by mutTF703/mutTR700 was198 bp in length.
Fig. 1.
Screening the optimal primers and validating probes for the detection of E. miricola. 2% agarose gel electrophoresis of RPA products produced by nine sets of primer combinations was performed (A). Availability of mutT1 probe by RPA-LFD assay (B). Positive: in the reaction system, E. miricola DNA sample was used as template, and probe was mutT1 probe. Negative: in the reaction system, H2O was used as template and probe was mutT1 probe. Blank: in the reaction system, no template was used and probe is mutT1 probe. Availability of mutT2 probe by exo RPA assay (C). Positive: in the reaction system, E. miricola DNA sample was used as template, and probe was mutT2 probe. Negative: in the reaction system, H2O was used as template and probe was mutT2 probe.
The verification of mutT1 probe and mutT2 probe based on primer pair mutTF703/mutTR700 yielded the positive results. The sample added with mutT1 probe and DNA sample showed specific band on dipstick in RPA-LFD assay, but the samples with water (negative) as template or only with mutT1 probe (blank) showed no specific bands (Fig. 1B). Compared with the negative control group, the sample added with mutT2 probe produced obvious fluorescence in exo RPA detection (Fig. 1C).
3.2. Specificities of basic RPA, RPA-LFD and exo RPA assays
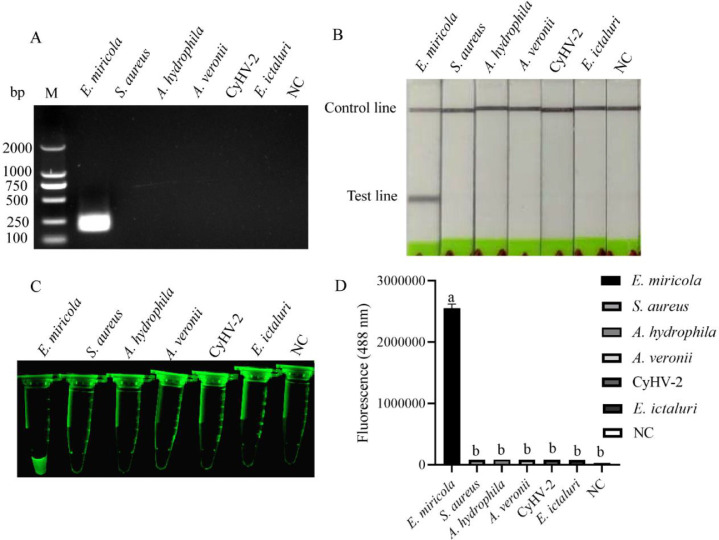
The specificities of RPA, RPA-LFD and exo RPA methods were verified by DNA samples of 6 pathogens. The E. miricola DNA obtained positive results by means of gel electrophoresis (Fig. 2A), lateral flow dipstick (Fig. 2B) and fluorescence imaging system (Fig. 2C), while S. aureus DNA, A. hydrophila DNA, A. veronii DNA, CyHV-2 DNA and E. ictalurid DNA did not. The fluorescence of exo RPA product was quantified by enzyme-labeled instrument. The results showed that fluorescence intensity value of E. miricola sample was significantly higher than that of others (Fig. 2D). These results suggest that RPA, RPA-LFD and exo RPA methods can specifically detect E. miricola.
Fig. 2.
Specificity detection of RPA, RPA-LFD and exo RPA. The samples for RPA,RPA-LFD and exo RPA specificity assays were different pathogens. RPA products were exhibited by agarose gel electrophoresis (A), lateral flow dipstick (B), fluorescence imaging system (C). The fluorescence intensities of exo RPA products were detected by the enzyme-labeled instrument (D). “NC” indicates negative control. (n = 3). Different lowercase letters indicate significant differences between groups.
3.3. Sensitivities of basic RPA, RPA-LFD and exo RPA assays
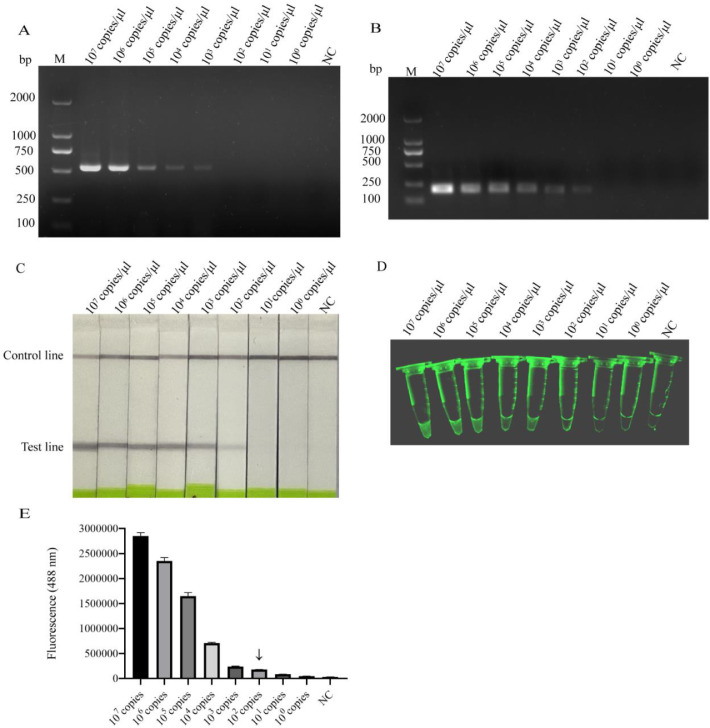
The 10°–107 copies/μL diluted standard plasmid sample of pMD19-mutT were detected by PCR, RPA, RPA-LFD and exo RPA. The results showed the minimum detectable concentration of generic PCR was 103 copies/μL (Fig. 3A), and that of RPA, RPA-LFD and exo RPA were 102 copies/μL (Fig. 3B-3D). The sensitivity of basic RPA was the same as that of RPA-LFD and exo RPA, which was ten-fold higher than that of conventional PCR. The fluorescence intensity values of exo RPA products were proportional to the concentration of standard plasmid in the range of 107–102 copies/μL. The limit of visible fluorescence of exo RPA was 102 copies/μL (Fig. 3E).
Fig. 3.
Sensitivities of PCR, RPA, RPA-LFD and exo RPA assays. The pMD19-mutT standard plasmid was diluted to 10°–107 copies/μL, and then used to detect minimum detectable concentration by PCR (A), RPA (B), RPA-LFD (C) and exo RPA (D). The fluorescence intensities of exo RPA products were detected by the enzyme-labeled instrument (E), the symbol ↓ represents the limit of visible fluorescence. “NC” indicates negative control. (n = 3).
3.4. Tissue distribution of E. miricola
The morphology and color of E. miricola were observed and determined by culturing E. miricola in BHI medium. This was used to distinguish E. miricola from commensal bacteria. We used healthy frog tissues for smearing plate in 5 replicates and did not detect E. miricola. However, E. miricola was detected in the samples of frogs with significant crooked head and cataract symptoms (Fig. S1).
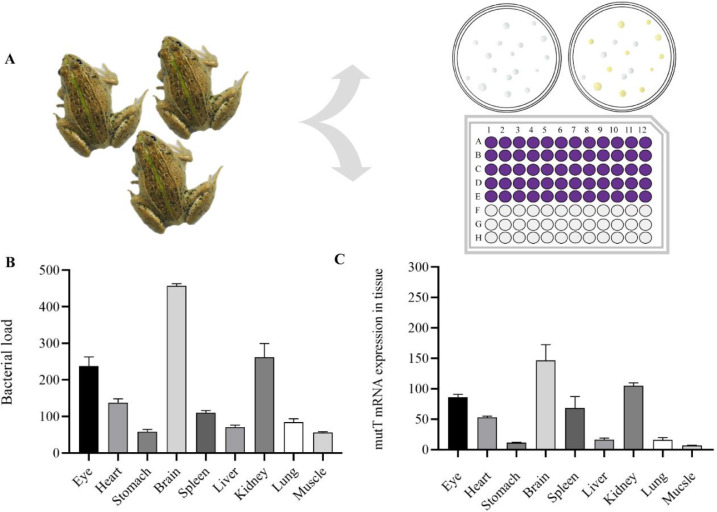
The tissues of five sick black spotted frogs were collected containing eye, heart, stomach, brain, spleen, liver, kidney, lung and muscle. The bacterial load of these tissues was analyzed by dilution coated plate method and the expression of E. miricola gene in tissues by qRT-PCR (Fig. 4A). Colony counts indicated that the brain loaded the most abundant bacteria (Fig. 4B). The results of qRT-PCR assay were consistent with the results of tissue bacterial load assay (Fig. 4C).
Fig. 4.
Screening tissues with high bacterial load. Pattern diagram of bacterial load detection methods (A). Tissue bacterial load was detected by dilution coated plate method (B), and the expression of mutT in tissues was detected by qRT-PCR (C).
3.5. Practicability of RPA, RPA-LFD and exo RPA assays
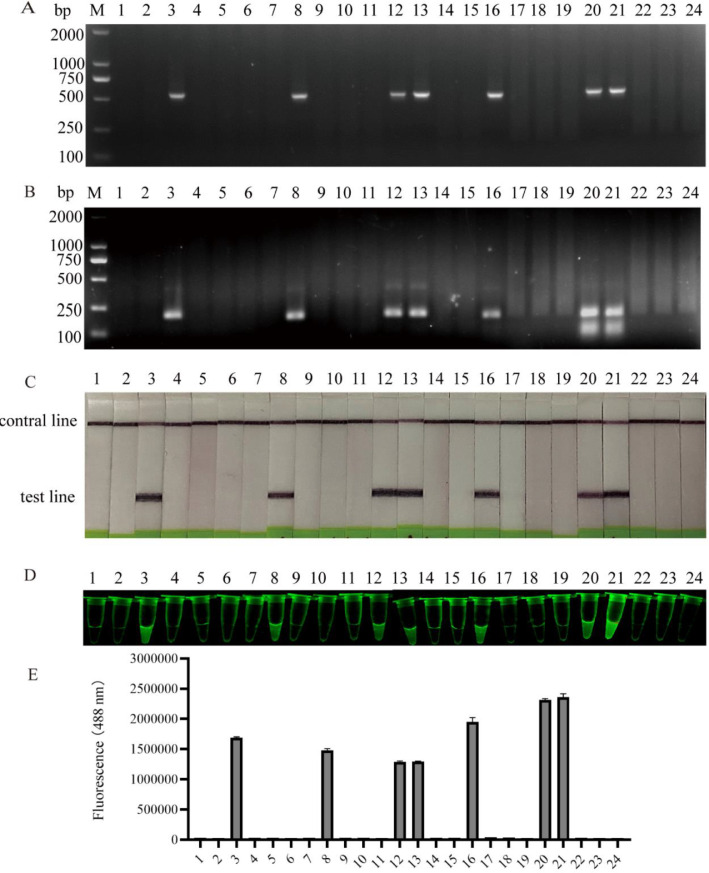
24 clinical samples were randomly selected for practical testing. The clinical performance of RPA, RPA-LFD and exo RPA was evaluated by comparing the PCR assay result of the same samples. The results showed that 7 positive samples were detected by the PCR assay (Fig. 5A), and 7 positive samples were also detected by RPA, RPA-LFD and exo RPA assays (Fig. 5B-5D). The four methods had the same detection positive rate. The fluorescence intensity values of exo RPA products detected by the enzyme-labeled instrument showed that only 7 samples had high fluorescence (Fig. 5E).
Fig. 5.
Evaluation of the clinical performance of PCR, RPA, RPA-LFD and exo RPA. Random 24 frog brain DNA were tested by conventional PCR (A), RPA (B), RPA-LFD (C) and exo RPA (D). The fluorescence intensity of the 24 exo RPA products were detected with an enzyme-labeled instrument (E).
4. Discussion
Facing the detriment of bacterial diseases with high infectivity and high mortality in frogs, developing low-cost, rapid and sensitive detection methods is a urgent and highly prominent problem [34]. Rapid and reliable diagnostic techniques play an important role in efficiently detecting E. miricola. In the present study, we developed the RPA-LFD and exo RPA methods for rapid detection of E. miricola. RPA-LFD and exo RPA, which have the advantages of simple operation, sensitive detection and low cost of production, were extensively used in fundamental research and industry [18,32,33,35,36]. However, the RPA method was rarely used in amphibians. We reported the application of RPA related technology in black spotted frogs for the first time.
Improper gene sequences selection can affect the specificity of RPA reaction [37]. In previous study, a real-time PCR detection system based on mutT gene sequence amplification was established, which can specifically identify E. miricola [12]. We used the mutT gene sequence as the target sequence to establish the detection system of RPA-LFD and exo RPA technologies, which detected the E. miricola specifically.
Probes of RPA-LFD and exo RPA are important components of the probe accuracy. The distance between the dT-fluorophore and dT-quenching group is an important factor in the exo RPA reaction, which affects the intensity of fluorescence. The distance between the two groups was 1–4 nt, and the optimum distance was 1–2 nt [29]. Fluorescence is a luminous phenomenon. According to previous reports, the maximum excitation and maximum emission wavelengths of FAM and SYBR Green are similar, and the fluorescence emission peaks are almost completely superimposed [38,39]. We used FAM channel to detect fluorescence. In future experiments, SYBR Green channel can also be used if conditions are limited, or a portable FAM machine (IT-IS UK) can be purchased, which is portable, lightweight and can work a whole day charging by battery.
Sensitivity and specificity assays are the basis to verify the practicability of RPA-LFD and exo RPA. In previous studies, the sensitivity of RPA-LFD and exo RPA methods are ten-fold higher than that of PCR, and the target bacteria DNA can be specifically detected in a variety of different pathogens [25,32]. In this study, the sensitivity of RPA-LFD and exo RPA is 102 copies/μL, which ten-fold higher than that of generic PCR (103 copies/μL), and E. miricola was specifically detected in different pathogens. These results showed that RPA-LFD and exo RPA are suitable methods for detecting pathogens and have potential application.
In order to sample quickly and accurately in clinical detection, we screened the tissues with high bacterial load in naturally diseased frogs by dilution coated plate method and qRT-PCR. The dilution coated plate method is a reliable method for calculating the bacterial load in tissues [40]. qRT-PCR is a classical method for detecting the expression of bacteria in tissues [31]. The results found that the brain was the main invading tissue of E. miricola. This is consistent with previous studies that the brain was the main target organ of E. miricola [5,7]. In addition, among the 24 clinical samples of black spotted frog, 7 samples were positive for E. miricola by RPA, RPA-LFD and exo RPA assays, which were also positive by the PCR generic assay. The positive rates of the five detection methods were the same.
5. Conclusion
RPA-LFD and exo RPA can achieve rapid (within 30 min), accurate (long primers and probes ensure the specificity), sensitive (102 copies/μL), convenient (without expensive equipment), practical detection of E. miricola, which contribute to the early diagnosis in frog culture farms. The results were analyzed in practical applications, and showed that RPA, RPA-LFD and exo RPA have an excellent potential for rapid diagnosis of E. miricola.
CRediT authorship contribution statement
Meihua Qiao: Conceptualization, Investigation, Writing – original draft. Liqiang Zhang: Visualization, Investigation. Jiao Chang: Methodology, Investigation, Validation. Haoxuan Li: Methodology, Investigation, Validation. Jingkang Li: Data curation, Writing – original draft. Weicheng Wang: Methodology, Investigation, Validation. Gailing Yuan: Methodology, Investigation, Validation. Jianguo Su: Conceptualization, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Mr. Xingchen Huo, Mr. Zhensheng Wang, Mr. Chuang Xu, Mr. Aotian Ouyang, Mr. Tianbing Xu, Mr. Bo Liang, Mr. Jie Zhang and Miss Yan He for technical advice and assistance in experiments. This work was supported by Innovative Project of Wuhan Academy of Agricultural Sciences (CXJSFW202103–2) and Fundamental Research Funds for the Central Universities (2662021SCPY006).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fsirep.2022.100059.
Appendix. Supplementary materials
References
- 1.Kim K.K., Kim M.K., Lim J.H., Park H.Y., Lee S.T. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int. J. Syst. Evol. Microbiol. 2005;55(3):1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- 2.Trimpert J., Eichhorn I., Vladimirova D., Haake A., Schink A.K., Klopfleisch R., Lubke-Becker A. Elizabethkingia miricola infection in multiple anuran species. Transbound Emerg. Dis. 2020;68(2):931–940. doi: 10.1111/tbed.13761. [DOI] [PubMed] [Google Scholar]
- 3.Elizabeth C., Isabel C., Carla F., Fernando C. Joint infection due to Elizabethkingia miricola. Span. J. Chemother. 2020;33(2):141–142. doi: 10.37201/req/081.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta P., Zaman K., Mohan B., Taneja N. Elizabethkingia miricola: a rare non-fermenter causing urinary tract infection. World J. Clin. Cases. 2017;5(5):187–190. doi: 10.12998/wjcc.v5.i5.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu R.X., Yuan J.F., Meng Y., Wang Z., Gu Z.M. Pathogenic Elizabethkingia miricola infection in cultured black-spotted frogs, China, 2016. Emerg. Infect. Dis. 2016;23(12):2055–2059. doi: 10.3201/eid2312.170942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei X.P., Yi G., Wang K.Y., OuYang P., Chen F., Huang X.L., Huang C., Lai W.M., Zhong Z.J., Huo C.L., Yang Z.X. Elizabethkingia miricola infection in Chinese spiny frog (Quasipaa spinosa) Transbound. Emerg. Dis. 2019;66(2):1049–1053. doi: 10.1111/tbed.13101. [DOI] [PubMed] [Google Scholar]
- 7.Huang X., Feng Y., Tang H., Xiong G., Li L., Yang Y., Wang K., Ouyang P., Geng Y., Chen D., Yang S.Y. Candidate animal disease model of Elizabethkingia spp. infection in humans, based on the systematic pathology and oxidative damage caused by E. miricola in Pelophylax nigromaculatus. Oxid. Med. Cell. Longev. 2019;2019:13. doi: 10.1155/2019/6407524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X.B., Liu L.Y., Tu Y.P., Zhang J., Miao G.J., Zhang L.L., Ge S.X., Xia N.S., Yu D.L., Qiu X.B. Rapid PCR powered by microfluidics: a quick review under the background of COVID-19 pandemic. Trends Anal. Chem. 2021;143 doi: 10.1016/j.trac.2021.116377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vantarakis A., Komninou G., Venieri D., Papapetropoulou M. Development of a multiplex PCR detection of Salmonella spp. and Shigella spp. in mussels. Lett. Appl. Microbiol. 2000;31(2):105–109. doi: 10.1046/j.1365-2672.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- 10.Phillips D.E., Mee P.T., Lynch S.E., da Conceicao F., Bendita da Costa Jong J., Rawlin G.T. Use of field based loop mediated isothermal amplification (LAMP) technology for a prevalence survey and proof of freedom survey for African swine fever in timor-leste in 2019. Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.672048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong X., Xu W.J., Guo L.Q., An J.X., Huang L.L., Qian H.Y., Cui X.W., Li Y., Cao M., Xiong X.H., Ying X.G., Wang L.B. Development of loop-mediated isothermal amplification (LAMP) assay for rapid screening of skipjack tuna (Katsuwonus pelamis) in processed fish products. J. Food Compos. Anal. 2021;102 [Google Scholar]
- 12.Zhang Q., Hu R.X., Gu Z.M. A real-time PCR assay for detection of emerging infectious Elizabethkingia miricola. Mol. Cell. Probes. 2020;52 doi: 10.1016/j.mcp.2020.101571. [DOI] [PubMed] [Google Scholar]
- 13.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Zhou S.T., Wen J.X., Sun M., Jiang Y.S., Lu L.Q., Xie J. A real-time reverse-transcription isothermal recombinase polymerase amplification assay for the rapid detection of genotype III grass carp (Ctenopharyngodon idella) reovirus. J. Virol. Methods. 2020;277 doi: 10.1016/j.jviromet.2019.113802. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z.W., Huang J., Zhang F., Zhou Y., Huang H.J. Detection of shrimp hemocyte iridescent virus by recombinase polymerase amplification assay. Mol. Cell. Probes. 2020;49 doi: 10.1016/j.mcp.2019.101475. [DOI] [PubMed] [Google Scholar]
- 16.Cong F., Zeng F.W., Wu M.L., Wang J.J., Huang B.H., Wang Y.Y., Wang Q., Zhang S.Q., Ma L., Guo P.J., Zeng W.W. Development of a real-time reverse transcription recombinase polymerase amplification assay for rapid detection of spring viremia of carp virus. Mol. Cell. Probes. 2020;50 doi: 10.1016/j.mcp.2019.101494. [DOI] [PubMed] [Google Scholar]
- 17.Li H.X., Yuan G.L., Luo Y.Z., Yu Y.Z., Ai T.S., Su J.G. Rapid and sensitive detection of infectious spleen and kidney necrosis virus by recombinase polymerase amplification combined with lateral flow dipsticks. Aquaculture. 2020;519 [Google Scholar]
- 18.Cabada M.M., Malaga J.L., Castellanos-Gonzalez A., Bagwell K.A., Naeger P.A., Rogers H.K., Maharsi S., Mbaka M., Clinton White J.A. Recombinase polymerase amplification compared to real-time polymerase chain reaction test for the detection of Fasciola hepatica in human stool. Am. J. Trop. Med. Hyg. 2017;96(2):341–346. doi: 10.4269/ajtmh.16-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao F.M., Zhang J.X., Li N., Chen T., Mi L.G., Zhang J.Y., Wang S.C., Wang Y., Zhang S.F., Hu R.L. Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African swine fever virus. Front. Microb. Immunol. 2019;10:1004. doi: 10.3389/fmicb.2019.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Sun M., Xu D., Podok P., Xie J., Jiang Y., Lu L. Rapid visual detection of cyprinid herpesvirus 2 by recombinase polymerase amplification combined with a lateral flow dipstick. J. Fish Dis. 2018;41:1201–1206. doi: 10.1111/jfd.12808. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y., Qin X.D., Zhang X.G., Zhao Z.X., Zhang W., Zhu X.L., Cong G.Z., Li Y.M., Zhang Z.D. Development of real-time and lateral flow dipstick recombinase polymerase amplification assays for rapid detection of goatpox virus and sheeppox virus. Virol. J. 2017;14(1):131. doi: 10.1186/s12985-017-0792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J.Q., Wang Y., Su H.J., Ding H.M., Sun X.C., Gao H., Geng Y., Wang Z.C. Rapid analysis of Escherichia coli O157:H7 using isothermal recombinase polymerase amplification combined with triple-labeled nucleotide probes. Mol. Cell. Probes. 2020;50 doi: 10.1016/j.mcp.2019.101501. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L.W., Wang J.C., Sun X.X., Wang J.F., Chen Z.M., Xu X.D., Dong M.Y., Guo Y.N., Wang Y.Y., Chen P.P., Gao W.J., Geng Y.Y. Development and evaluation of the rapid and sensitive RPA assays for specific detection of salmonella spp. in food samples. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.631921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava N., Kapoor R., Kumar R., Kumar S., K S.R., Kumar S., Baranwal V.K. Rapid diagnosis of Cucumber mosaic virus in banana plants using a fluorescence-based real-time isothermal reverse transcription-recombinase polymerase amplification assay. J. Virol. Methods. 2019;270:52–58. doi: 10.1016/j.jviromet.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y., Qin X.D., Wang G.X., Zhang Y.E., Shang Y.J., Zhang Z.D. Development of a fluorescent probe-based recombinase polymerase amplification assay for rapid detection of Orf virus. Virol. J. 2015;12:206. doi: 10.1186/s12985-015-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J.P., Liu X.Q., Chen J., Guo Z.Y., Wang Y.Y., Chen G.P., Chen X.J., Yan Q.Y., Yang P., Li R.S., Yang G.W., Lan Q.X., Wang J.L. Development of a rapid test method for Salmonella enterica detection based on fluorescence probe-based recombinase polymerase amplification. Food Anal. Methods. 2019;12(8):1791–1798. [Google Scholar]
- 27.Han Q.Y., Wang J.F., Li R.W., Han Q.G., Yuan W.Z., Wang J.C. Development of a recombinase polymerase amplification assay for rapid detection of Haemophilus parasuis in tissue samples. Vet. Med. Sci. 2020;6(4):894–900. doi: 10.1002/vms3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia X.M., Yu Y.X., Weidmann M., Pan Y.J., Yan S.L., Wang Y.J. Rapid detection of shrimp white spot syndrome virus by real time, isothermal recombinase polymerase amplification assay. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0104667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murinda S.E., Ibekwe A.M., Zulkaffly S., Cruz A., Park S., Razak N., Paudzai F.M., Ab Samad L., Baquir K., Muthaiyah K., Santiago B., Rusli A., Balkcom S. Real-time isothermal detection of shiga toxin-producing Escherichia coli using recombinase polymerase amplification. Foodborne Pathog. Dis. 2014;11(7):529–536. doi: 10.1089/fpd.2013.1663. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y.Q., Xiao X., Hu Y.Z., Liao Z.W., Zhu W.T., Jiang R., Yang C.R., Zhang Y.A., Su J.G. CXCL20a, a teleost-specific chemokine that orchestrates direct bactericidal, chemotactic, and phagocytosis-killing-promoting functions, contributes to clearance of bacterial infections. J. Immunol. 2021;207(7):1911–1925. doi: 10.4049/jimmunol.2100300. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z.S., Huo X.C., Zhang Y.Q., Gao Y., Su J.G. Carboxymethyl chitosan nanoparticles loaded with bioactive protein CiCXCL20a effectively prevent bacterial disease in grass carp (Ctenopharyngodon idella) Aquaculture. 2022;549 [Google Scholar]
- 32.Li H.X., Zhang L.Q., Yu Y.Z., Zhang Y.A., Su J.G. Rapid detection of Edwardsiella ictaluri in yellow catfish (Pelteobagrus fulvidraco) by real-time RPA and RPA-LFD. Aquaculture. 2022;552 [Google Scholar]
- 33.Liu X.Q., Yan Q.Y., Huang J.F., Chen J., Guo Z.Y., Liu Z.D., Cai L., Li R.S., Wang Y., Yang G.W., Lan Q.X. Influence of design probe and sequence mismatches on the efficiency of fluorescent RPA. World J. Microbiol. Biotechnol. 2019;35(6):95. doi: 10.1007/s11274-019-2620-2. [DOI] [PubMed] [Google Scholar]
- 34.Rahim G.R., Gupta N. Elizabethkingia miricola: discrepancies in identification and antimicrobial susceptibilities. Diagn. Microbiol. Infect. Dis. 2019;94(1):104. doi: 10.1016/j.diagmicrobio.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z.Y., Wang Y., Lin L., Wu T., Zhao Z.Z., Ying B.W., Chang L.Q. A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113663. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y., Qin P.Z., He J., Li W.W., Shi Y.L., Xu J.G., Wu Q., Chen Q.Q., Li W.D., Wang X.X., Liu G.D., Chen W. Rapid and simultaneous visual screening of SARS-CoV-2 and influenza virufses with customized isothermal amplification integrated lateral flow strip. Biosens. Bioelectron. 2022;197 doi: 10.1016/j.bios.2021.113771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daher R.K., Stewart G., Boissinot M., Boudreau D.K., Bergeron M.G. Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol. Cell. Probes. 2015;29(2):116–121. doi: 10.1016/j.mcp.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 38.He S.J., Song B., Li D., Zhu C.F., Qi W.P., Wen Y.Q., Wang L.H., Song S.P., Fang H.P., Fan C.H. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010;20(3):453–459. [Google Scholar]
- 39.Poucke M.V., Zeveren A.V., Peelman L.J. Combined FAM-labeled TaqMan probe detection and SYBR green I melting curve analysis in multiprobe qPCR genotyping assays. BioTechniques. 2012;52(2):81–86. doi: 10.2144/000113808. [DOI] [PubMed] [Google Scholar]
- 40.Xu C., Qiao M.H., Huo X.C., Liao Z.W., Su J.G. An oral microencapsulated vaccine loaded by sodium alginate effectively enhances protection against GCRV infection in grass carp (Ctenopharyngodon idella) Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.848958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.