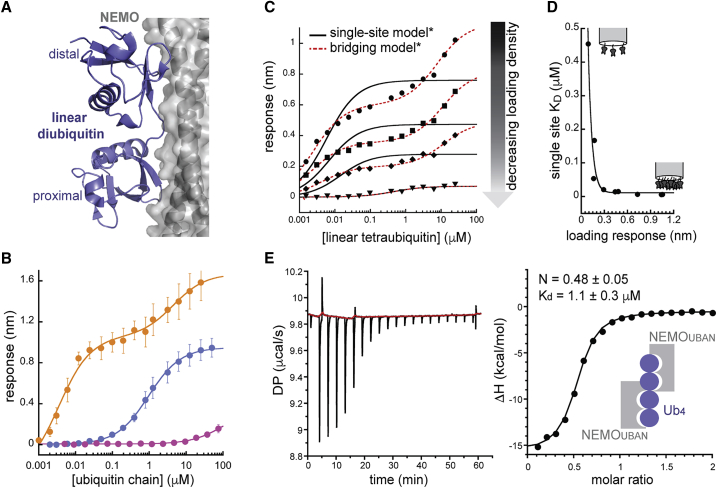
Figure 2.
Ubiquitin binding by NEMOUBAN. (A) NEMOUBAN (gray cartoon and surface) selectively recognizes linear diubiquitin (blue cartoon) as a single binding unit (Protein Data Bank (PDB): 2ZVO). (B) BLI measurements of monobiotinylated NEMOUBAN binding to discrete ubiquitin species. Error bars represent standard deviations from three measurements. Fits to single-site binding model (mono- and diubiquitin) or bridging model (tetraubiquitin) shown. For visual clarity, the data here are fit with Rmin set to zero (see Figs. S1, c and d for a comprehensive analysis of single-site and bridging model fits, including comparisons of fitting models with and without Rmin as a fitted parameter) (C) Linear tetraubiquitin binding singly biotinylated NEMOUBAN at various loading densities: 1.0 nm (circles), 0.73 nm (squares), 0.44 nm (diamonds), and 0.09 nm (triangles) loading response. Fits to single-site binding equation or bridging model shown. (D) Apparent KD-values derived from single-site model change as a function of loading density. Data fit to an exponential decay equation to guide the eye. (E) ITC analysis of NEMOUBAN binding to linear tetraubiquitin. A representative titration is shown both as raw injection heats over the titration time course (left panel) and as integrated and normalized heats per titration step (right panel), with the latter fit to a single-site-binding model (solid line). Average KD and complex stoichiometry values are reported with standard deviations (n = 3) and support a 2:1 model of NEMOUBAN/tetraubiquitin binding. All fitting parameters from fits of these BLI data may be found in Table S1.